
The B cell receptor in marginal zone lymphoma ontogeny and evolution
Introduction
The 2016 Revision of the World Health Organization (WHO) Classification of Lymphoid Neoplasms recognizes three distinct entities of marginal zone lymphomas (MZL): (I) extranodal MZL of mucosa-associated lymphoid tissue (MALT lymphoma), (II) nodal MZL; and, (III) splenic MZL (1-3). These entities differ in terms of pathophysiology, tissues involved, genetic features, clinical presentation, prognosis and possible treatments (4). However, a common feature shared by most if not all MZL concerns a pronounced role of interactions with the microenvironment and selection by (auto)antigens throughout their natural history (5). This places the B cell receptor immunoglobulin (BcR IG) expressed by the malignant clones at the frontlight, given its critical role as mediator of antigen-specific drive affecting the ontogeny and evolution of MZL.
Immunogenetic studies in MZL have offered insight into the intricate relationship between the respective malignant clones and their surrounding microenvironment, through revealing biases in the IG gene repertoire and particular imprints of somatic hypermutation (SHM) in the context of (ongoing) antigen selection, while also highlighting associations with chronic inflammation and/or analogies with polyreactive B cells (5,6). This is the focus of the present review.
B cell receptors in lymphomas—key to improved understanding, key to refined management
Mature B cell lymphomas arise from well differentiated B cells: unsurprisingly, therefore, the malignant B cells retain many features of their normal counterparts, including engagement in multifaceted interactions with the microenvironment. That said, the nature of such interactions and relative contribution to lymphomagenesis varies considerably between different lymphomas (7,8).
The tumor microenvironment of B cell lymphomas is a complex network consisting of heterogeneous cell populations (including macrophages, follicular reticular cells, follicular dendritic cells, fibroblasts, endothelial cells and T cells, amongst others) as well as soluble mediators e.g., chemokines, cytokines and adhesion molecules, to name a few (7,9,10). Lymphoma cells engage in reciprocal interactions with accessory cells, their mediators and structural components of the extracellular matrix (11,12): for such interactions to occur, the malignant B cells critically rely on communication pathways whereby signals are sensed via appropriate receptors.
Similar to normal B cells, the malignant cells in mature B cell lymphomas express a diverse repertoire of surface receptors that transmit and augment or, in other instances, dampen microenvironmental signals, eventually affecting cell behavior: of these, the most critical role is played by the B cell receptor immunoglobulin (BcR IG), a claim supported by various lines of evidence (6).
First, it is highly pertinent that B cell lymphomas exhibit disease-restricted IG gene repertoires and particular imprints of SHM within the clonotypic BcR IG, strongly suggestive of functional selection by discrete antigens or classes of related antigenic elements (6,13). Second, most lymphoma cells do not survive or proliferate autonomously ex vivo, highlighting a critical dependence on microenvironmental triggers (7,14). Third, inhibition of microenvironmental crosstalk with pharmacological agents directly targeting relevant signaling pathways, including those originating from the BcR IG, has emerged as a highly efficacious novel therapeutic paradigm for various B cell lymphomas (15). Finally, recurrent genomic aberrations in these entities frequently cluster in B cell pathways critical for microenvironmental interactions, including the BcR, Toll-like receptor (TLR), Notch, NF-κB and MAPK pathways, indicating functional selection (16).
Microenvironmental interactions in MZL
The three distinct subtypes of MZL (extranodal of MALT type, nodal, splenic) have specific diagnostic criteria and disease-biased biology, clinical presentation and natural history (4). The situation is not as straightforward when it comes to provisional entities in the 2016 WHO Classification, such as splenic leukemia/lymphoma unclassifiable (SLLU) (17,18), while it becomes more complicated by the recent recognition of clonal lymphocytosis of marginal zone origin (CBL-MZ) (19). This term refers to the presence of circulating clonal B cells with an immunophenotype suggestive of marginal zone derivation in individuals with no symptoms or signs of lymphoma. CBL-MZ is still enigmatic and may in fact represent an ‘umbrella’ covering several, unrelated conditions: that said, evidence suggests that CBL-MZ may precede the onset of overt MZL (especially SMZL), therefore constituting a pre-malignant condition. Notably, there is not a defined cut-off in the clonal B-cell lymphocyte count for discriminating CBL-MZ from MZL (19,20).
As their name implies, MZ lymphomas appear to arise in the defined microanatomical area defined as the MZ. Normally, the MZ hosts different types of B cells, including monocytoid B cells and MZ cells. The precise ontogenetic relationship of monocytoid and MZ cells is still disputed, with some authors claiming that both belong to the same B cell subpopulation (21), contrasting others who favor distinct origins (22). In support of the latter, monocytoid B cells have been shown to be sIgM-/BCL2- cells mostly expressing BcR IG with unmutated IGHV genes, whereas MZ B cells are sIgM+/BCL2+ cells generally exhibiting low levels of SHM (22,23). Moreover, monocytoid B cells are located in the marginal zones adjacent to the subcapsular and intermediary sinuses, whereas MZ B cells are found in different anatomical sites albeit most characteristically in the spleen, where they are located at the boundary between the red and the white pulp (24-26). At this location, they are constantly exposed to blood-borne pathogens and constitute a first-line of defense against T cell-independent antigens, such as polysaccharide antigens found on encapsulated bacteria, while they may also participate in T cell-dependent immune responses to protein as well as lipid antigens. Hence, MZ cells are considered as a bridge between innate and adaptive immune responses, and they are thought to be endowed with “natural memory” (25-27).
MZ cells are a major source of natural antibodies present in healthy individuals in the absence of exogenous antigenic stimulation: these are predominantly IgM, and exhibit polyreactivity against self and exogenous antigens (28). The repertoire and reactivity pattern of natural antibodies is stable both within and across species, perhaps reflecting the usage of specific germline-encoded antigen specificities (i.e. IG genes) selected over evolution for their ability to recognize a wide range of antigens (29). Thus, MZ B cells not only provide protection against pathogens, but also serve important housekeeping roles, including the recognition and removal of senescent cells and other cellular debris (30).
For MZ cells to fulfil their role, communication with the microenvironment is essential, as they need to interact with different constituents of the innate and adaptive immune systems in order to initiate rapid antibody responses (27). Continuous (auto)antigenic stimulation of the BcR IG can lead to proliferation of B cells, while recruitment of inflammatory cells (such as T cells, neutrophils and macrophages) and other microenvironmental components contributes to lymphomagenesis (31), promoting B cell expansion and eventual transformation.
Microenvironmental dependencies are maintained in the distinct MZL entities. Despite the differences between extranodal MALT lymphomas, NZML and SMZL, they appear to have in common the key implication of (auto)antigenic stimulation and microenvironmental interactions in their ontogeny. In fact, MZLs are considered to represent a paradigmatic example of external triggering in lymphoma development, with environmental factors and the host immune response contributing significantly to their pathogenesis (32). This is supported by different lines of evidence, most importantly the connection between different MZL entities and chronic inflammation in the context of infections or autoimmune disorders (33).
The most representative example concerns gastric MALT lymphoma and infection by Helicobacter pylori (H. pylori) (34). Under physiological conditions, lymphocytes cannot survive in the gastric mucosa, due to the low pH of the stomach. However, when H. pylori infect the gastric mucosa they secrete urease buffering the pH of the stomach, allowing infiltration by lymphocytes and gradual acquisition of MALT (31,35,36), thus creating a permissive microenvironment for B cell clonal expansion and gastric MALT lymphoma outgrowth.
In general, chronic inflammation emerges as a universal feature in the development of MALT lymphomas, as it leads to the accumulation of B cells in organs that usually lack lymphoid tissue (stomach, lung, salivary glands or ocular adnexa) (31). Besides infection by H. pylori in gastric MALT lymphoma, a similar scenario has been described for Chlamydophila psittaci (Cp) in ocular adnexal MALT lymphoma (OAMZL) (37,38); Borrelia burgdorferi in cutaneous MALT lymphoma (39); Campylobacter jejuni in immunoproliferative small intestinal disease (32,40); and, the hepatitis C virus in SMZL and extranodal MALT lymphomas (41-43). It should not be forgotten, however, that extranodal MZL may arise in the context of chronic inflammation associated with autoimmune disorders e.g., Sjögren’s syndrome in the case of MALT lymphoma of the salivary glands and Hashimoto’s thyroiditis in the case of MALT lymphoma of the thyroid (33,44).
Immunoglobulin gene repertoire restrictions in MZL
Immunogenetic studies have been key in unravelling biases in the IG gene repertoire in all three distinct MZL entities (Figure 1), suggesting that antigens or superantigens are implicated in lymphomagenesis through stimulating the proliferation of B cells with particular IG receptors.

Extranodal MZL
Extranodal MALT lymphomas display restricted usage of the IGHV1-69 gene, albeit with differential distribution depending on the primary site of involvement: indeed, this gene is more frequently expressed in salivary MALT lymphoma, followed by gastric MALT and OAMZL (45). Other IG gene repertoire biases described for extranodal MALT lymphomas concern (I) frequent usage of the IGHV3-7 gene in gastric MALT lymphoma and OAMZL (45); (II) restricted pairing of IGHV4-34 and IGHV1-8 gene rearrangements with IGKV3-20 gene rearrangements (50).
Immunogenetic studies have also supported antigen selection in the natural history of extranodal MALT lymphomas also by revealing strong similarities to antibodies with rheumatoid factor (RF) reactivity. In more detail, elegant studies from van Noesel’s group have revealed striking VH CDR3 homology with RFs in almost 20% of gastric MALT lymphomas and 41% of salivary gland MALT lymphomas and, through in vitro binding studies, provided functional evidence that a significant fraction of such cases display strong RF reactivity (51). Moreover, their studies have identified that stereotypic RFs are frequently expressed in MALT lymphomas of the salivary glands but are rare in the labial salivary glands of patients with Sjögren’s syndrome, again strongly supporting selection (52).
Along the same lines, another particularly interesting case is presented by OAMZL, which accounts for ~15% of all extranodal MALT lymphomas and ~80% of lymphomas arising in the ocular adnexa (conjunctiva, orbit, eyelid, and lachrymal gland and sac) (38). OAMZL has been shown to be significantly associated, albeit with variable geographic prevalence, with infection by Chlamydophila psittaci (Cp), the cause of human psittacosis (37,40,53). Immunogenetic studies by us and others have reported that certain IGHV genes predominate in the repertoire of OAMZL, again with variable frequency depending on the geographical area. Indicatively, in a series of 44 cases, mostly from Europe, we showed that only two IGHV genes, namely IGHV3-23 and IGHV3-7, acounted for 26% of all cases (54). Of note, Cp-negative cases have been shown to display a different IG gene repertoire, with overuse of IGHV4-34, perhaps reflecting distinct antigen exposure histories from Cp-positive OAMZL (55).
In our study of the BcR IG repertoire in OAMZL 5/44 cases (11%) carried BcR IG with high homology to antibodies from autoreactive clones; all 5 cases were Cp-positive (54). This has prompted us to argue for selection by restricted autoantigenic elements and propose a scenario whereby clonotypic BcR IG in OAMZL progenitors but also the malignant B cells themselves may be triggered by autoantigenic stimulation. According to this scenario, pertinent for other if not all extranodal MZL as well, though through other implicated antigenic triggers, Cp leads to the development of an inflammatory milieu, where autoantigens are exposed and presented, hence promoting the selection and proliferation of autoreactive B cell clones. If a genetic aberration occurs in an intensely proliferating clone, this would provide the necessary drive for the emergence of an uncontrolled B cell expansion and, eventually, OAMZL.
Nodal MZL
IGHV biases have also been reported in different studies of NMZL, with the IGHV1-69 and IGHV4-34 genes predominating (45,47). However, the investigated series were small, precluding firm conclusions from being drawn.
Splenic MZL
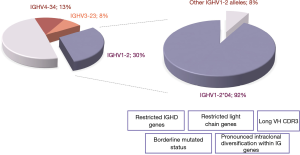
Amongst all MZLs, perhaps the most impressive repertoire skewing has been documented in SMZL. In fact, close to half of SMZL cases express one of only three genes, namely IGHV1-2, IGHV4-34 and IGHV3-23, with clear predominance of the former that accounts for roughly one-third of SMZL (48,49,56-58). Noticeably, the IGHV1-2 gene was under-represented among cases with spleen histopathology suggestive of splenic diffuse red pulp lymphoma (SDRL), one of the major provisional entities in the SLLU category (49). This finding highlights its ontogenetic distance from SMZL, while also implying that distinct selection processes may underlie the development of SMZL and SDRL.
A notable feature of the IG repertoire in SMZL concerns the fact that restrictions in gene usage extend to the level of polymorphic variants i.e. alleles of IGHV genes. In fact, the vast majority (~90%) of IGHV1-2 expressing SMZL cases utilize allele *04, a single polymorphic variant of the IGHV1-2 gene (49). This bias is quite remarkable, considering that the frequency of this allele is much lower in other B cell lymphomas (59,60). IGHV1-2*04 cases also display a series of distinctive immunogenetic features, setting them apart from SMZL cases utilizing other IGHV genes (49). In particular, these cases carry unususally long VH CDR3, particularly compared to the remaining SMZL (22 amino acids vs 16 amino acids in non-IGHV1-2*04 cases); biased IGHD gene usage (over-representation of the IGHD3-3, IGHD3-9 and IGHD3-10 genes), leading to the presence of IGHD-encoded restricted peptide motifs at the tip of the VH CDR3; and, restricted pairings with certain light chain genes (IGKV3-20, IGKV1-8 and IGLV2-14) (61).
Allele *04 of the IGHV1-2 gene has unique immunogenetic properties since it encodes for a tryptophan (W) residue at VH FR3 position 75, whereas other alleles of both the IGHV1-2 and almost all other IGHV genes in all species examined thus far carry an arginine (R) residue at this position (62). The presence of R-75 appears to be critical for stabilizing VH FR3 and the overall IG structure, through the formation of an ion bond with a highly conserved aspartic acid (D) residue located at VH FR3 position 98 of all functional IGHV genes (62,63). Hence, what could be the reason for the remarkable selection of this peculiar *04 allele, appearing to represent an ‘anomaly of nature’?
We have previously reported that the presence of W-75 could have an important impact on BcR IG conformation. In fact, through molecular dynamics simulation we provided evidence that the R-to-W replacement at position VH FR3-75 could result in a substantial alteration in antibody conformation, with apparent closure of the VH CDR3 cavity. These in silico findings indicate that W-75 likely has an important structural role, and allude to the recognition of specific antigenic epitopes through the conferred structural conformation (49,64). The nature of such epitopes and the importance of the dominance of this germline-encoded specificity in SMZL remain enigmatic, not least due to the scant published evidence (65).
That notwithstanding, it would not be unreasonable to argue that these immunogenetic findings point to the existence of a distinct molecular variant of SMZL defined by usage of IGHV1-2*04 with implications for distinct ancestry and/or distinct immune activation processes from SMZL cases utilizing other IGHV genes. This might imply that SMZL has multiple cells of origin, with SMZL ontogeny perhaps reflecting the heterogeneity of B cell populations residing within the normal splenic MZ, including few naïve B cells, follicular cells as well as MZ cells, which we have previously proposed as the postulated counterparts of IGHV1-2*04 SMZL (49,64).
Somatic hypermutation of the BcR IG in MZL: biological and clinical implications
The key role of the clonotypic BcR IG in MZL pathogenesis is underscored by the fact that the great majority of cases bear imprints of SHM. Indicatively, in a systematic immunogenetic study by our group (45), only a small fraction of cases was assigned to the ‘truly unmutated’ subgroup (i.e. cases with IGHV genes lacking any SHM and displaying 100% identity to the germline) ranging from 6.2% in extranodal MALT lymphoma (6/98 cases) to 8.2% in NMZL (3/37 cases) and 12.2% in SMZL (43/353 cases). The ‘borderline/minimally mutated’ subgroup (97–99.9% IGHV gene germline identity) accounted for 18.3% of extranodal MALT lymphoma (18/98 cases), 18.9% of NMZL (7/37 cases) and 37.4% of SMZL (132/353 cases). Finally, 50.4% of SMZL cases (178/353), 72.9% of the NMZL cases (27/37) and 75.5% of the MALT lymphoma cases (74/98) were classified as ‘significantly mutated’ i.e had a germline identity below 97% due to carrying a high number of SHMs (45).
Particularly noteworthy profiles of SHM have been identified in IGHV1-2*04 SMZL, where the vast majority of cases are classified as ‘borderline/minimally mutated’. Intriguingly, SHMs reported in IGHV1-2*04 BcR IG are often recurrent, cluster in framework regions (FRs) and are conservative in nature (i.e. lead to substitutions by amino acids with similar physicochemical properties as the germline-encoded amino acid) (49,61) (Figure 2). Altogether, the aforementioned immunogenetic signature strongly implicates selection pressure by antigen(s) in the outgrowth of IGHV1-2*04 clones. This claim gained ground when it was shown that IGHV1-2*04 cases also display pronounced intraclonal diversification within the IG genes, far exceeding cases utilizing other IGHV genes. This finding has prompted the argument that SHM in IGHV1-2*04 cases is an ongoing process continuously fine-tuning the structure of the clonotypic BcR IG under pressure by antigens (66).
Similar results were obtained from independent studies in both NMZL and MALT lymphomas. In more detail, Granai et al. (47), reported that 20/28 NMZL cases (71.5%) were mutated, with only 2/28 (7%) of the unmutated cases displaying 100% GI. Finally, in the study by Craig et al. (67), 82% of the gastric MALT lymphoma cases (9/11) belonged to the IG-mutated subgroup.
IGHV gene SHM status in MZL: does it have a prognostic value?
Given the precedent of CLL where the SHM status of the clonotypic rearranged IGHV genes holds great value as a prognostic/predictive biomarker (68,69), it is not paradoxical that the question arose as to whether this biomarker could be clinically relevant also in MZLs. Summarizing the published evidence, the actual impact of the IGHV gene SHM status on the clinical course of MZL remains rather equivocal, with independent studies reaching different conclusions.
Particularly for SMZL, where the evidence base is more solid, Rinaldi et al. (70) analyzed 83 SMZL cases, 51 of which were mutated (61%) and 32 unmutated (39%), following the 98% germline identity cut-off value. The adoption of this ‘CLL-relevant’ cut-off raises concerns, since the majority of SMZL cases indeed carry some level of SHM, characterized by “disease-specific” patterns (49). In simple words, the application of the 98% germline identity cut-off leads to a biologically questionable assumption in that all sequences with IGHV gene SHM status above the applied cut-off are defined as unmutated even when carrying one or more mutations (<100% germline identity) (6). That reservation notwithstanding, no statistically significant differences were identified between IG-mutated versus IG-unmutated cases concerning clinical features and outcome (70). A similar result was obtained in a subsequent study (71) of 55 SMZL patients of whom 14 (25%) were IG-unmutated while 41 (75%) were IG-mutated (again, following the 98% cut-off), where, again, no statistically significant differences were identified in either overall survival or progression-free survival between the two IG mutational subgroups. In contrast, in a more recent study by our group, we have reported that the absence of any trace of SHM (‘truly unmutated’ IGHV genes) was an independent marker of short time-to-first-treatment (72). Hence, the jury is still out regarding the potential clinical implications of IGHV gene SHM status in SMZL, if any.
Moving to NMZL, Granai et al. (47) reported significant differences between patients with mutated (20/28 cases, 71.5%) versus unmutated (8/28 cases, 28.5%) BcR IG in terms of disease-specific survival and progression-free survival. However, these differences did not extend to overall survival. The clinical impact of SHM has also been tested in extranodal MALT lymphomas, with studies showing no clinicopathological differences between IG-mutated versus IG-unmutated cases (73).
Altogether, the study of the potential clinical impact of SHM has not led to definitive conclusions. This could be attributed, at least in part, to the small patient cohorts analyzed and the heterogeneous treatments administered, introducing significant biases, thus clearly pointing to the need for more extensive studies.
Distinctive landscapes of cell-intrinsic aberrations for distinctive immunogenetic subgroups of MZL
A common characteristic of MZL concerns the presence of recurrent chromosomal and/or genomic aberrations targeting B cell pathways that are crucial for microenvironmental interactions (16). Against that similarity, however, the genetic background of MZL subtypes is characterized by distinct profiles (Figure 3).
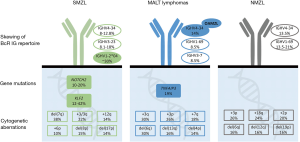
Regarding cytogenetics, a comprehensive study of 330 SMZL cases revealed that the majority (238/330; 72%) had an abnormal karyotype (77). Concerning individual genomic aberrations, del(7q) was found to predominate (42/114 cases, 38%) (78), followed by gains of 3/3q (17/76 cases, 22%), 12q (11/76 cases, 14%), 6p (8/76 cases, 10%), and deletions of 8p (12/76 cases, 15%), 17p (14/76 cases, 14%) and 13q (9/76 cases, 12%) (78,79). Numerical chromosomal aberrations have also been reported in extranodal MALT lymphomas, with, for example, gains in 3q, 3p and 7q and losses of 6q, 13q and 4p (80). In NMZL, some of the most common genomic aberrations are gains in 3p, 18q and 2p, and deletions in 6q, 12q and 19p (80). It should be noted, however, that for both extranodal MALT lymphomas and NMZL, the respective series were small, thus additional investigation is clearly warranted.
Structural aberrations, particularly translocations, are rare in SMZL and NMZL, thus contrasting extranodal MALT lymphomas, where 4 recurrent translocations have been reported, namely t(11,18)(q21;q21)/API2-MALT1, t(14,18)(q32;q21)/IGH-MALT1, t(1,14)(p22;q32)/BCL10-IGH (81), and t(3,14)(p13;q32)/FOXP1- IGH (82), with varying incidence depending on the site of the MALT lymphoma (83-85).
Turning to gene mutations, the list of genes frequently mutated in MZL is quite extensive. That said, asymmetries have been noted between different MZL (Figure 3): for example, SMZL is characterized by frequent mutations in the NOTCH2 (72,86) and KLF2 (72,74,75) genes (with reported incidences in the range of 10–20% and 12–42%, respectively), that are far less frequent in NMZL or extranodal MALT lymphomas (74,75,86). Importantly, many of these mutated genes are involved in crucial B cell signalling pathways, including the NF-κB pathway (both the canonical and the non-canonical), leading to the constitutive activation of NF-κB (87). This further highlights the importance of external drive in MZL natural history.
Besides genomic aberrations, epigenetic modifications such as DNA methylation can also have an impact on the evolution of MZL (88). For instance, studies in SMZL have revealed an overall higher level of promoter methylation in cases with stronger propensity for histologic transformation and worse survival. In particular, the cases with high promoter methylation (33/134 cases, 25%) were enriched for del(7q), NOTCH2 mutations and expression of the IGHV1-2*04 gene (76). This finding is in line with the results of independent studies revealing asymmetric distribution of genomic aberrations in IGHV1-2*04 versus other immunogenetic subgroups of SMZL: in more detail, a strong correlation was observed between mutations in KLF2 gene and IGHV1-2*04 expression (72).
On the evidence presented above, it appears that in SMZL a distinctive immunogenetic signature can be associated with a distinctive landscape of cell-intrinsic aberrations (Figure 3). More recent findings indicate that this pattern may also be found in other MZL as well, most notably in OAMZL, where a remarkable enrichment of mutations in the TNFAIP3 gene was reported in cases expressing the IGHV4-34 gene versus other IGHV genes. This finding implies a synergistic effect between BcR IG carrying the IGHV4-34 gene and TNFAIP3 mutations in the ontogenesis of OAMZL. In more specific, in a study of 63 patients with OAMZL, the majority of IGHV4-34 cases carried a mutation in TNFAIP3 (7/10 cases, 70%) (46). According to this line of reasoning, chronic activation of the IGHV4-34 BcR may induce pronounced B cell activation, eventually favouring the genetic inactivation of TNFAIP3, a key regulator of inflammation signalling pathways and a negative regulator of the NF-κB pathway. TNFAIP3 inactivation would, thus, lead to overactive NF-κB signalling independently of the presence of extrinsic triggers, setting the stage for the development of OAMZL as the final outcome of this particular cell-extrinsic/cell-intrinsic interaction (89).
Concluding remarks
BcR IG gene repertoire profiling has been key for understanding MZL, offering compelling evidence that their natural history is influenced by external drive, thus pointing to antigen-experienced cells as the respective progenitor cells for all MZL distinct entities. The importance of antigenic stimulation is further supported by the association between BcR IG with distinct immunogenetic properties and specific infectious agents and/or autoimmune diseases, as well as the enrichment of certain cell-intrinsic aberrations in MZL subgroups with distinctive BcR IG.
On these grounds, our final statement could not be other than that the BcR IG is the critical driver in MZL ontogeny and evolution.
Acknowledgments
Funding: This work was supported in part by COSMIC, a Marie Curie European Training Network funded from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 765158; NOVEL, an ERANET Transcan-2 project; CLLon, funded by the Hellenic Foundation for Research and Innovation and GSRT, the General Secretariat for Research and Technology of Greece (CLLon grant agreement no. 336); and the Hellenic Precision Medicine Network in Oncology.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Francesco Bertoni, Thomas Habermann, Davide Rossi, Emanuele Zucca) for the series “Marginal Zone Lymphomas” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-20-18). The series “Marginal Zone Lymphomas” was commissioned by the editorial office without any funding or sponsorship. KS has received unrestricted research support from Janssen. MP is honoraria from Janssen-Cilag, Novartis, Genesis-Pharma and Alexion. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Isaacson PG, Du MQ. MALT lymphoma: From morphology to molecules. Nat Rev Cancer 2004;4:644-53. [Crossref] [PubMed]
- Campo E, Swerdlow SH, Harris NL, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood 2011;117:5019-32. [Crossref] [PubMed]
- Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375-90. [Crossref] [PubMed]
- Zucca E, Arcaini L, Buske C, et al. Marginal zone lymphomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:17-29. [Crossref] [PubMed]
- Gemenetzi K, Agathangelidis A, Zaragoza-Infante L, et al. B Cell Receptor Immunogenetics in B Cell Lymphomas: Immunoglobulin Genes as Key to Ontogeny and Clinical Decision Making. Front Oncol 2020;10:67. [Crossref] [PubMed]
- Sutton LA, Agathangelidis A, Belessi C, et al. Antigen selection in B-cell lymphomas-Tracing the evidence. Semin Cancer Biol 2013;23:399-409. [Crossref] [PubMed]
- Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer 2014;14:517-34. [Crossref] [PubMed]
- Mulder TA, Wahlin BE, Österborg A, et al. Targeting the Immune Microenvironment in Lymphomas of B-Cell Origin: From Biology to Clinical Application. Cancers (Basel) 2019;11:915. [Crossref] [PubMed]
- Herreros B, Sanchez-Aguilera A, Piris MA. Lymphoma microenvironment: Culprit or innocent? Leukemia 2008;22:49-58. [Crossref] [PubMed]
- Coupland SE. The challenge of the microenvironment in B-cell lymphomas. Histopathology 2011;58:69-80. [Crossref] [PubMed]
- Palucka AK, Coussens LM. The Basis of Oncoimmunology. Cell 2016;164:1233-47. [Crossref] [PubMed]
- Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012;21:309-22. [Crossref] [PubMed]
- Stevenson FK, Sahota SS, Ottensmeier CH, et al. The occurrence and significance of V gene mutations in B cell-derived human malignancy. Adv Cancer Res 2001;83:81-116. [Crossref] [PubMed]
- Young RM, Wu T, Schmitz R, et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci U S A 2015;112:13447-54. [Crossref] [PubMed]
- Jerkeman M, Hallek M, Dreyling M, et al. Targeting of B-cell receptor signalling in B-cell malignancies. J Intern Med 2017;282:415-28. [Crossref] [PubMed]
- Rossi D, Ciardullo C, Gaidano G. Genetic aberrations of signaling pathways in lymphomagenesis: Revelations from next generation sequencing studies. Semin Cancer Biol 2013;23:422-30. [Crossref] [PubMed]
- Matutes E. Immunophenotyping and Differential Diagnosis of Hairy Cell Leukemia. Hematol Oncol Clin North Am 2006;20:1051-63. [Crossref] [PubMed]
- Traverse-Glehen A, Baseggio L, Callet-Bauchu E, et al. Splenic red pulp lymphoma with numerous basophilic villous lymphocytes: A distinct clinicopathologic and molecular entity? Blood 2008;111:2253-60. [Crossref] [PubMed]
- Xochelli A, Kalpadakis C, Gardiner A, et al. Clonal B-cell lymphocytosis exhibiting immunophenotypic features consistent with a marginal-zone origin: Is this a distinct entity? Blood 2014;123:1199-206. [Crossref] [PubMed]
- Xochelli A, Oscier D, Stamatopoulos K. Clonal B-cell lymphocytosis of marginal zone origin. Best Pract Res Clin Haematol 2017;30:77-83. [Crossref] [PubMed]
- Tierens A, Delabie J, Michiels L, et al. Marginal-zone B cells in the human lymph node and spleen show somatic hypermutations and display clonal expansion. Blood 1999;93:226-34. [Crossref] [PubMed]
- Stein K, Hummel M, Korbjuhn P, et al. Monocytoid B cells are distinct from splenic marginal zone cells and commonly derive from unmutated naive B cells and less frequently from postgerminal center B cells by polyclonal transformation. Blood 1999;94:2800-8. [Crossref] [PubMed]
- Lazzi S, Bellan C, Tiacci E, et al. IRTAI+ monocytoid B cells in reactive lymphadenitis show a unique topographic distribution and immunophenotype and a peculiar usage and mutational pattern of IgVH genes. J Pathol 2006;209:56-66. [Crossref] [PubMed]
- Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol 2002;2:323-35. [Crossref] [PubMed]
- Pillai S, Cariappa A, Moran ST. Marginal Zone B Cells. Annu Rev Immunol 2005;23:161-96. [Crossref] [PubMed]
- Weill JC, Weller S, Reynaud CA. Human Marginal Zone B Cells. Annu Rev Immunol 2009;27:267-85. [Crossref] [PubMed]
- Cerutti A, Cols M, Puga I. Marginal zone B cells: Virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol 2013;13:118-32. [Crossref] [PubMed]
- Baumgarth N, Tung JW, Herzenberg LA. Inherent specificities in natural antibodies: A key to immune defense against pathogen invasion. Springer Semin Immunopathol 2005;26:347-62. [Crossref] [PubMed]
- Kirkham PM, Schroeder HW Jr. Antibody structure and the evolution of immunoglobulin V gene segments. Semin Immunol 1994;6:347-60. [Crossref] [PubMed]
- Lopes-Carvalho T, Kearney JF. Development and selection of marginal zone B cells. Immunol Rev 2004;197:192-205. [Crossref] [PubMed]
- Thieblemont C, Bertoni F, Copie-Bergman C, et al. Chronic inflammation and extra-nodal marginal-zone lymphomas of MALT-type. Semin Cancer Biol 2014;24:33-42. [Crossref] [PubMed]
- Ferreri AJM, Govi S, Ponzoni M. Marginal zone lymphomas and infectious agents. Semin Cancer Biol 2013;23:431-40. [Crossref] [PubMed]
- Bende RJ, Van Maldegem F, Van Noesel CJM. Chronic inflammatory disease, lymphoid tissue neogenesis and extranodal marginal zone B-cell lymphomas. Haematologica 2009;94:1109-23. [Crossref] [PubMed]
- Marcelis L, Tousseyn T, Sagaert X. MALT lymphoma as a model of chronic inflammation-induced gastric tumor development. Curr Top Microbiol Immunol 2019;421:77-106. [Crossref] [PubMed]
- Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, et al. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 1991;338:1175-6. [Crossref] [PubMed]
- Zucca E, Bertoni F, Roggero E, et al. Molecular analysis of the progression from Helicobacter pylori-associated chronic gastritis to mucosa-associated lymphoid-tissue lymphoma of the stomach. N Engl J Med 1998;338:804-10. [Crossref] [PubMed]
- Ferreri AJM, Guidoboni M, Ponzoni M, et al. Evidence for an association between Chlamydia psittaci and ocular adnexal lymphomas. J Natl Cancer Inst 2004;96:586-94. [Crossref] [PubMed]
- Sassone M, Ponzoni M, Ferreri AJM. Ocular adnexal marginal zone lymphoma: Clinical presentation, pathogenesis, diagnosis, prognosis, and treatment. Best Pract Res Clin Haematol 2017;30:118-30. [Crossref] [PubMed]
- Garbe C, Stein H, Dienemann D, et al. Borrelia burgdorferi—associated cutaneous B cell lymphoma: Clinical and immunohistologic characterization of four cases. J Am Acad Dermatol 1991;24:584-90. [Crossref] [PubMed]
- Ponzoni M, Ferreri AJM. Bacteria associated with marginal zone lymphomas. Best Pract Res Clin Haematol 2017;30:32-40. [Crossref] [PubMed]
- Armand M, Besson C, Hermine O, et al. Hepatitis C virus - Associated marginal zone lymphoma. Best Pract Res Clin Haematol 2017;30:41-9. [Crossref] [PubMed]
- Xiong W, Lv R, Li H, et al. Prevalence of hepatitis B and hepatitis C viral infections in various subtypes of B-cell non-Hodgkin lymphoma: confirmation of the association with splenic marginal zone lymphoma. Blood Cancer J 2017;7:e548 [Crossref] [PubMed]
- Defrancesco I, Zerbi C, Rattotti S, et al. HCV infection and non-Hodgkin lymphomas: an evolving story. Clin Exp Med 2020;20:321-8. [Crossref] [PubMed]
- Bertoni F, Rossi D, Zucca E. Recent advances in understanding the biology of marginal zone lymphoma. F1000Res 2018;7:406. [Crossref] [PubMed]
- Xochelli A, Bikos V, Polychronidou E, et al. Disease-biased and shared characteristics of the immunoglobulin gene repertoires in marginal zone B cell lymphoproliferations. J Pathol 2019;247:416-21. [Crossref] [PubMed]
- Moody S, Escudero-Ibarz L, Wang M, et al. Significant association between TNFAIP3 inactivation and biased immunoglobulin heavy chain variable region 4-34 usage in mucosa-associated lymphoid tissue lymphoma. J Pathol 2017;243:3-8. [Crossref] [PubMed]
- Granai M, Amato T, Di Napoli A, et al. IGHV mutational status of nodal marginal zone lymphoma by NGS reveals distinct pathogenic pathways with different prognostic implications. Virchows Arch 2020;477:143-50. [Crossref] [PubMed]
- Zibellini S, Capello D, Forconi F, et al. Stereotyped patterns of B-cell receptor in splenic marginal zone lymphoma. Haematologica 2010;95:1792-6. [Crossref] [PubMed]
- Bikos V, Darzentas N, Hadzidimitriou A, et al. Over 30% of patients with splenic marginal zone lymphoma express the same immunoglobulin heavy variable gene: Ontogenetic implications. Leukemia 2012;26:1638-46. [Crossref] [PubMed]
- Zhu D, Lossos C, Chapman-Fredricks JR, et al. Biased immunoglobulin light chain use in the Chlamydophila psittaci negative ocular adnexal marginal zone lymphomas. Am J Hematol 2013;88:379-84. [Crossref] [PubMed]
- Bende RJ, Aarts WM, Riedl RG, et al. Among B cell non-Hodgkin’s lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J Exp Med 2005;201:1229-41. [Crossref] [PubMed]
- Bende RJ, Slot LM, Hoogeboom R, et al. Stereotypic rheumatoid factors that are frequently expressed in mucosa‐associated lymphoid tissue-type lymphomas are rare in the labial salivary glands of patients with Sjögren’s syndrome. Arthritis Rheumatol 2015;67:1074-83. [Crossref] [PubMed]
- Chanudet E, Zhou Y, Bacon CM, et al. Chlamydia psittaci is variably associated with ocular adnexal MALT lymphoma in different geographical regions. J Pathol 2006;209:344-51. [Crossref] [PubMed]
- Dagklis A, Ponzoni M, Govi S, et al. Immunoglobulin gene repertoire in ocular adnexal lymphomas: Hints on the nature of the antigenic stimulation. Leukemia 2012;26:814-21. [Crossref] [PubMed]
- van Maldegem F, Wormhoudt TAM, Mulder MMS, et al. Chlamydia psittaci-negative ocular adnexal marginal zone B-cell lymphomas have biased v H 4-34 immunoglobulin gene expression and proliferate in a distinct inflammatory environment. Leukemia 2012;26:1647-53. [Crossref] [PubMed]
- Stamatopoulos K, Belessi C, Papadaki T, et al. Immunoglobulin heavy-and light-chain repertoire in splenic marginal zone lymphoma. Mol Med 2004;10:89-95. [Crossref] [PubMed]
- Traverse-Glehen A, Davi F, Ben Simon E, et al. Analysis of VH genes in marginal zone lymphoma reveals marked heterogeneity between splenic and nodal tumors and suggests the existence of clonal selection. Haematologica 2005;90:470-8. [PubMed]
- Hockley SL, Giannouli S, Morilla A, et al. Insight into the molecular pathogenesis of hairy cell leukaemia, hairy cell leukaemia variant and splenic marginal zone lymphoma, provided by the analysis of their IGH rearrangements and somatic hypermutation patterns. Br J Haematol 2010;148:666-9. [Crossref] [PubMed]
- Agathangelidis A, Darzentas N, Hadzidimitriou A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: A molecular classification with implications for targeted therapies. Blood 2012;119:4467-75. [Crossref] [PubMed]
- Hadzidimitriou A, Agathangelidis A, Darzentas N, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood 2011;118:3088-95. [Crossref] [PubMed]
- Bikos V, Stalika E, Baliakas P, et al. Selection of antigen receptors in splenic marginal-zone lymphoma: Further support from the analysis of the immunoglobulin light-chain gene repertoire. Leukemia 2012;26:2567-9. [Crossref] [PubMed]
- Lefranc M. The global ImMunoGeneTics Web Resource for immunoglobulins (IG), T cell receptors (TR) and major histocompatibility (MH) and related proteins of the immune system (RPI) 2020. Available online: http://www.imgt.org/IMGTrepertoire/.
- Pommié C, Levadoux S, Sabatier R, et al. IMGT standardized criteria for statistical analysis of immunoglobulin V‐REGION amino acid properties. J Mol Recognit 2004;17:17-32. [Crossref] [PubMed]
- Baliakas P, Strefford JC, Bikos V, et al. Splenic marginal-zone lymphoma: Ontogeny and genetics. Leuk Lymphoma 2015;56:301-10. [Crossref] [PubMed]
- Warsame AA, Aasheim HC, Nustad K, et al. Splenic marginal zone lymphoma with VH1-02 gene rearrangement expresses poly- and self-reactive antibodies with similar reactivity. Blood 2011;118:3331-9. [Crossref] [PubMed]
- Bikos V, Karypidou M, Stalika E, et al. An immunogenetic signature of ongoing antigen interactions in splenic marginal zone lymphoma expressing IGHV1-2*04 receptors. Clin Cancer Res 2016;22:2032-40. [Crossref] [PubMed]
- Craig VJ, Arnold I, Gerke C, et al. Gastric MALT lymphoma B cells express polyreactive, somatically mutated immunoglobulins. Blood 2010;115:581-91. [Crossref] [PubMed]
- Rosenquist R, Ghia P, Hadzidimitriou A, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: Updated ERIC recommendations. Leukemia 2017;31:1477-81. [Crossref] [PubMed]
- Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018;131:2745-60. [Crossref] [PubMed]
- Rinaldi A, Forconi F, Arcaini L, et al. Immunogenetics features and genomic lesions in splenic marginal zone lymphoma. Br J Haematol 2010;151:435-9. [Crossref] [PubMed]
- Arcaini L, Zibellini S, Passamonti F, et al. Splenic marginal zone lymphoma: Clinical clustering of immunoglobulin heavy chain repertoires. Blood Cells Mol Dis 2009;42:286-91. [Crossref] [PubMed]
- Parry M, Rose-Zerilli MJJ, Ljungström V, et al. Genetics and prognostication in splenic marginal zone lymphoma: Revelations from deep sequencing. Clin Cancer Res 2015;21:4174-83. [Crossref] [PubMed]
- Yoshida M, Okabe M, Eimoto T, et al. Immunoglobulin VH genes in thymic MALT lymphoma are biased toward a restricted repertoire and are frequently unmutated. J Pathol 2006;208:415-22. [Crossref] [PubMed]
- Clipson A, Wang M, De Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia 2015;29:1177-85. [Crossref] [PubMed]
- Piva R, Deaglio S, Famà R, et al. The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia 2015;29:503-7. [Crossref] [PubMed]
- Arribas AJ, Rinaldi A, Mensah AA, et al. DNA methylation profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood 2015;125:1922-31. [Crossref] [PubMed]
- Salido M, Baró C, Oscier D, et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: A multicenter study of the Splenic B-Cell Lymphoma Group. Blood 2010;116:1479-88. [Crossref] [PubMed]
- Fresquet V, Robles EF, Parker A, et al. High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br J Haematol 2012;158:712-26. [Crossref] [PubMed]
- Watkins AJ, Hamoudi RA, Zeng N, et al. An integrated genomic and expression analysis of 7q deletion in splenic marginal zone lymphoma. PLoS One 2012;7:e44997 [Crossref] [PubMed]
- Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood 2011;117:1595-604. [Crossref] [PubMed]
- Streubel B, Simonitsch-Klupp I, Müllauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004;18:1722-6. [Crossref] [PubMed]
- Streubel B, Vinatzer U, Lamprecht A, et al. T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005;19:652-8. [Crossref] [PubMed]
- Liu H, Ye H, Ruskone-Fourmestraux A, et al. T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 2002;122:1286-94. [Crossref] [PubMed]
- Du MQ. MALT lymphoma: recent advances in aetiology and molecular genetics. J Clin Exp Hematop 2007;47:31-42. [Crossref] [PubMed]
- Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia 2010;24:483-7. [Crossref] [PubMed]
- Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med 2012;209:1537-51. [Crossref] [PubMed]
- Rossi D, Deaglio S, Dominguez-Sola D, et al. Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood 2011;118:4930-4. [Crossref] [PubMed]
- Casulo C, Friedberg J. Transformation of marginal zone lymphoma (and association with other lymphomas). Best Pract Res Clin Haematol 2017;30:131-8. [Crossref] [PubMed]
- Agathangelidis A, Xochelli A, Stamatopoulos K. A gene is known by the company it keeps: enrichment of TNFAIP3 gene aberrations in MALT lymphomas expressing IGHV4-34 antigen receptors. J Pathol 2017;243:403-6. [Crossref] [PubMed]
Cite this article as: Zaragoza-Infante L, Agathangelidis A, Papaioannou M, Chatzidimitriou A, Stamatopoulos K. The B cell receptor in marginal zone lymphoma ontogeny and evolution. Ann Lymphoma 2020;4:10.


