
Tailored treatment in mantle cell lymphoma
Introduction
Mantle cell lymphoma (MCL) is a non-eradicable lymphoproliferative disorder. Despite therapeutic successes achieved with the introduction of rituximab and high-dose regimens with cytarabine and autologous stem cell transplantation (ASCT), there is no survival plateau, and the course of the disease is characterized by a pattern of continuous relapses. However, MCL is a very heterogeneous disease; though the large majority of MCL patients achieve durable remission, some patients are refractory to treatment, with a worsening of outcome (1-5). Therefore, biological factors that influence the behavior of MCL and its response to treatment need to be identified. Clinical and biological factors have been identified and prognostic scores established, allowing the stratification of patients into risk classes. Although useful, these tools are not able to effectively identify very high or very low risk patients and cannot be applied for tailoring treatment (3). In recent decades, research has focused on identifying biological factors that affect the different clinical outcomes, such as DNA mutations (6) and specific mRNA signatures (7,8) identified by gene expression profiling (GEP) and protein expression (immunohistochemistry) (9). Moreover, minimal residual disease (MRD) analysis is an established tool in MCL; similar to other lymphoproliferative disorders, it allows us to assess the risk of recurrence during and after treatment by monitoring the residual malignant clone (5,10,11). In this review, we briefly discuss the validated tools for risk stratification in MCL and subsequently focus on the most appealing novel candidate biomarkers more extensively. Finally, we will discuss the potential application of this knowledge for tailored treatment in MCL patients.
Validated tools for risk stratification in mantle cell lymphoma
MIPI score
Given the inadequacy of the International Prognostic Index (IPI) in MCL, a dedicated prognostic tool was introduced into clinical practice in 2008, the MCL International Prognostic Index (MIPI) score (1). The MIPI score is based on independent clinical and laboratory prognostic factors: age, performance status according to the Eastern Cooperative Oncology Group (ECOG), lactate dehydrogenase (LDH), and leukocyte count. These parameters identified three groups of patients with different overall survival (OS): low risk (44% of patients, median OS not reached), intermediate risk (35%, 51 months OS), and high risk (21%, 29 months OS). This tool was subsequently validated in the context of modern rituximab and high-dose cytarabine clinical trials, the “MCL Younger” (12) and “MCL Elderly” (13) trials of the European MCL Network, in which the MIPI score identified three risk groups with 5-year OS rates of 83%, 63%, and 34% (low, intermediate, and high risk groups, respectively). Its impact was independent of treatment received and was valid for both younger and elderly patients. Moreover, high concordance was found between MIPI scores and the simplified MIPI (s-MIPI) score, which was more easily applied in clinical routine (2).
Ki-67 proliferative index
The histopathological marker Ki-67 is an indirect index of cell proliferation, identifying patients with more aggressive disease and worse prognosis (14-16). Attempting to improve the prognostic impact of the MIPI score, Ki-67 was integrated, giving rise to the “biological” MIPI (MIPI-b) (1). This validated tool was able to better identify patients with high-risk disease, but did not significantly discriminate between patients at low vs. intermediate risk (2). Finally, an improved model using the Ki-67 index as a dichotomous value (≥30%) was developed, the MIPI-c, which is able to identify four risk groups with 5-year OS of 85%, 72%, 43%, and 17% (3).
Blastoid morphology
The undifferentiated cytological aspect of MCL was identified as an unfavorable feature for both progression-free survival (PFS) and OS, regardless of the therapeutic progress with rituximab, high dose schedules, or novel combinations (3,17). Accordingly, the blastoid variant is more often associated with unfavorable clinical, immunohistochemical, and cytogenetic features, such as high MIPI score and Ki-67 index, and aberrations in TP53. ASCT is a valid option for young patients, but a combined approach with novel agents may be necessary to overcome the poor prognosis in elderly patients (18-21).
MRD analysis
MRD is defined as the small amount of disease that remains after an effective treatment, and is not identifiable by traditional imaging or laboratory techniques. The negative prognostic impact of MRD persistence measured by standardized allele-specific oligonucleotide (ASO) quantitative polymerase chain reaction (qPCR) has been validated in large phase II and III clinical trials (4,5,10,11,22).
Novel candidate biomarkers
Somatic mutations and chromosomal imbalances
In the last couple of years, the mutational landscape of MCL has been investigated extensively. The first whole genome and exome sequencing studies describing the characteristics and frequency of somatic mutations in MCL (6) grouped them on the basis of the physiological function impaired by the single gene aberration: genes controlling the cell cycle or responsible for DNA repair (CCND1, TP53, ATM), epigenetic regulation (KMT2D, WHSC1), and genes controlling cell-signaling pathways (NOTCH1/2, BIRC3, TRAF2). The most frequent mutations were recorded in ATM (41%), CCND1 (34%), TP53 (27%), KMT2D (13%), and WHSC1 (13%). However, the patient series described in these studies were retrospective, inhomogeneous, and not fully annotated, often leading to inconclusive results in terms of clinical impact. Some signals suggest a role of TP53, which is involved in the regulation of apoptosis and genomic stability and is altered in many hematological and solid tumors (23-25), and aberrations in NOTCH1/2, which encodes a single-pass transmembrane receptor and is prognostic in chronic lymphocytic leukemia (26), in the negative effects of mutations. More recently, in a highly selected unicentric series, MYC translocations were associated with particularly dismal outcomes (27).
Interestingly, both topographical and temporal heterogeneity were described in the mutational landscape of single MCL patients. In particular, different mutations were identified in different tissues collected at diagnosis (i.e., peripheral blood vs. lymph node), probably originating from a common precursor clone before spatially diverging with different tropism. Different clusters of mutations were also observed over time in the same patients, in accordance with clonal evolution of MCL cells between diagnosis and relapse (6).
More recent studies have focused on select mutations, analyzing their impact on outcome in prospective patient series (Table 1). The European MCL Network investigated somatic gene copy number alterations (CNAs) in 135 patients enrolled in the randomized “MCL Younger MCL” trial (#NCT00209222) (28). In this study, deletions in TP53 (22%) and CDKN2A [25%, encoding both the CDK4/6 inhibitor INK4a, p16, and the positive TP53 regulator ARF, p14 (30)] significantly impacted both PFS and OS. Moreover, the association of these two deletions (7%) conferred further worsening of the outcome, suggesting a synergistic negative effect.
Table 1
| Clinical trial | Patients (analyzed/enrolled) and age | Therapeutic schedule | Tissue | Technique | Candidate gene aberrations | Median PFS (pooled arms) | Median OS (pooled arms) |
|---|---|---|---|---|---|---|---|
| MCL Younger (NCT00209222) Ph III EuMCLNet trial (28) | 135/497, ≤65 years old | 6RCHOP vs. 3RCHOP/3RDHAP + ASCT | BM; PB; FFPE sections | Copy number variation analysis | del CDKN2A | CDKN2A: del 1.5 years vs. WT 5.8 years | CDKN2A: del 2.6 years vs. WT 6.8 years |
| Del TP53 | TP53: del 2.1 years vs. WT 5.8 years | TP53: del 4.1 years vs. WT 7 years | |||||
| MCL2 (ISRCTN 87866680); MCL3 (NCT00514475) NLG ph II trial (19) | 183/320, ≤65 years old | MCL2: MaxiCHOP/HDAraC + ASCT; MCL3: RmaxiCHOP/HDAraC ± Zevalin + ASCT | Unselected BM cells | NGS targeted resequencing | Mut TP53 | TP53: mut 0.9 years vs. WT 10.2 years | TP53: mut 1.8 years vs. WT NR |
| MCL0208 (NCT02354313) ph III FIL trial (21) | 186/300, ≤65 years old | RCHOP + HDAraC + ASCT ± lenalidomide maintenance | CD19-selected BM cells | NGS targeted resequencing/copy number variation analysis | Mut KMT2D | (4 year-PFS) KMT2D: mut 33% vs. WT 64% | (4 year-OS) KMT2D: mut 62% vs. WT 87% |
| Mut/Del TP53 | TP53: mut/del 25% vs. WT 63% | TP53: mut/del 55% vs. WT 88% | |||||
| MCL4 (NCT00963534) ph II NLG trial (29) | 46/50, ≥65 years old or ≤65 frail | Lenalidomide-bendamustine-rituximab | Unselected BM and PB cells | NGS targeted sequencing | Mut TP53 | TP53: mut 0.8 years vs. WT 3.5 years | TP53: mut 2.1 years vs. WT 5.8 years |
EuMCLNet, European Mantle Cell Lymphoma Network; NLG, Nordic Lymphoma Group; FIL, Fondazione Italiana Linfomi; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone; DHAP, cisplatinum, Ara-C, dexamethasone; ASCT, autologous stem cell transplantation; HDAraC, high-dose AraC; Lena-BeRit, lenalidomide, bendamustina, rituximab; BM, bone marrow; PB, peripheral blood; FFPE, formalin-fixed, paraffin-embedded; NGS, next-generation sequencing; Mut, mutation; Del, deletion; WT, wild type; mPFS, median progression-free survival; mOS, median overall survival; CDKN2A, cyclin-dependent kinase inhibitor 2A; TP53, tumor protein 53; KMT2D, histone-lysine N-methyltransferase 2D.
The negative impact of TP53 mutations in younger MCL patients was demonstrated in a combined prospective series from the “MCL2” and “MCL3” phase II trials of the Nordic Lymphoma Group (19), with mutated cases presenting a median OS of 1.8 years compared to not reached (NR) in wild type (WT) patients. In accordance with the European MCL Network study (28), deletions of TP53 and CDKN2A negatively affected prognosis, but a strong association with TP53 mutation was found in these cases.
Finally, the negative role of TP53 alterations (both mutations and deletions) was independently validated in the Italian series of the “MCL0208” trial by Fondazione Italiana Linfomi (FIL) (21). In this study, mutations in the gene encoding lysine methyltransferase 2D (KMT2D, known as MLL2), an epigenetic regulating enzyme that acts as a tumor suppressor, also independently predicted worse PFS and OS; these results were reproduced in the Nordic series (19) when more stringent bioinformatics criteria for KMT2D mutation calling were applied (21). On the other hand, NOTCH1 mutations were not confirmed as independent prognostic markers, as they often co-occurred with TP53 aberrations.
In summary, despite the increasing bulk of genomic data, the only validated biomarkers in MCL, thus far, are TP53 aberrations (both mutations and deletions) and, to a lesser extent, KMT2D mutations, both of which account for a median OS of 4 years in younger patients (Table 1). This impact is independent from the other known prognostic factors but partially associated with blastoid morphology, Ki67 ≥30%, and high-risk MIPI (19,21). Therefore, by adding KMT2D mutations and TP53 disruption to the MIPI-c backbone, the authors proposed a new genetic prognostic index, the “MIPI-g”, which improved the model discrimination ability compared to the MIPI-c alone (21). Moreover, no impact of the different treatment approaches was observed in young patients receiving high-dose therapy with ASCT.
However, despite the strong prognostic value, some limitations have to be clarified before the broad introduction of TP53 and KMT2D investigation to clinical routine. In particular, the tissue and analytical technique are heterogeneous, and the clinical impact in elderly patients has not yet been investigated in large prospective series (Table 1). These issues are currently being addressed in the context of the ongoing phase III clinical trials of the European MCL Network (e.g., “EuMCL-R2”, EudraCT 2012-002542-20 and “Triangle”, EudraCT 2014-001363-12). Nonetheless, these biomarkers are able to consistently select a population (~25% of patients) with a biologically different high-risk disease. These patients do not seem to benefit from the standard, high-dose chemo-immunotherapy, the current standard of care in young patients.
Immunohistochemistry
Immunohistochemical staining can be used to study the expression of some proteins in tissue for the diagnosis and characterization of MCL. In particular, increased expression of SOX11, a neural transcription factor not expressed in normal lymphoid tissue, is found in most MCL cases and in some cases of hairy cell leukemia (31), and it seems to be implicated in the regulation of cell differentiation (32). In small retrospective studies, the absence of SOX11 has been associated with a subgroup of patients with better prognosis, characterized by leukemic and splenic, but not nodal, disease (33), and frequently with hypermutated IGHV genes and low genome complexity (34), so-called “indolent MCL”. Recent studies with larger cohorts of patients have had contradictory results regarding the prognostic significance of this immunohistochemical marker. In Nordic studies, SOX11 expression was negatively associated with Ki-67 and p53 expression, identifying a population with low proliferative index and non-blastoid morphology, which is suggestive of it being a somewhat protective factor (25,35). In the cohort of patients from the European MCL Network trials, the presence of SOX11 did not correlate with time to treatment failure (TTF) or OS in a multivariate analysis with MIPI and Ki-67 (9). Therefore, data about this marker are still contradictory, and a possible explanation may be underrepresentation of indolent MCL cases in clinical trials.
On the other hand, the expression of p53 is considered a surrogate for the mutational status of TP53 (36). For a long time, increased expression of p53 has been associated with aggressive MCL. In large clinical trials from both the Nordic group and the European MCL Network, expression of p53 was significantly associated with poor outcomes (9,25). In particular, a high level of expression (>50% of cells) was strongly predictive of short TTF and OS in both univariate and multivariate analyses. In addition, most patients with high p53 expression have a high Ki-67 and high-risk MIPI. An epiphenomenon of p53 aberration may be a higher proliferative index and more aggressive clinical behavior. Moreover, Aukema et al. reported that a lack of p53 expression is associated with worse outcome, which may be a consequence of TP53 mutation, deletion, or epigenetic alterations negatively affecting the protein function. In summary, as the clinical value of p53 expression has been validated in numerous large prospective trials, independently of MIPI and Ki-67, incorporation of p53 staining into routine diagnostic practice is now recommended (37). However, its use is not yet widespread, and strict assessment guidelines need to be followed to ensure inter-laboratory reproducibility (38).
GEP
GEP has mostly been used in the last 20 years to improve the characterization of lymphoproliferative diseases, targeting both the malignant cell and the tumor microenvironment, mainly in diffuse large B-cell lymphoma (39-42) and follicular lymphoma (43,44). More recently, GEP has been applied to MCL studies in order to develop new stratification risk models. The main features of currently available GEP-based tools for risk stratification in MCL are summarized in Table 2.
Table 2
| Clinical trial/ reference | Tissue | Technology | Genes studied | Median PFS (pooled arms) | Median OS (pooled arms) |
|---|---|---|---|---|---|
| NCT00114738 (7) | 55 PB/FFPE samples | GEP | 27-gene BCR signature; 18-gene NF-kB signature; 28-gene NIK signature | BCR≥upper tercile 2 years; BCR<upper tercile 2.9 years | (7.5-year OS) BCR≥upper tercile 68%, BCR<upper tercile 96% |
| MCL0208 (NCT02354313) (45) | 83 PB/FFPE samples | GEP/qRT-PCR | 6-gene signature | BCRhigh 3.5 years; BCRlow not reached | NA |
| Retrospective series (8) | 110 FFPE samples | NanoString (MCL-35 assay) | 17-gene proliferation signature | NA | High risk 1.1 years; standard risk 2.6 years; low risk 8.6 years |
| MCL “Younger” (NCT00209222); MCL “Elderly” (NCT00209209) (46) | 169 FFPE samples | NanoString (MCL-35 assay) | 17-gene proliferation signature | High risk 0.7 years; standard risk 2.6 years; low risk 5.3 years | “Younger”: high risk 0.8 years, standard risk 4.7 years, low risk 10 years; “Elderly”: high risk 2 years, standard risk 3 years, low risk NR |
| MCL2 (ISRCTN 87866680); MCL3 (NTC 00514475) (47) | 74 FFPE samples | NanoString (MCL-35 assay) | 17-gene proliferation signature; 18-housekeeping genes | High risk 2.8 years; standard risk NR; low risk 6.5 years | High risk 5 years; standard/low risk NR |
| Retrospective series (48) | 70 PB samples | NanoString (L-MCL16 assay) | 16-gene proliferation signature | (3-year TTT); nnMCL 31%; cMCL 88% | (3-year OS) nnMCL 92%; cMCL 69% |
FIL, Fondazione Italiana Linfomi; EuMCLNet, European MCL Network; NLG, Nordic Lymphoma Group; GEP, gene expression profile; PB, peripheral blood; FFPE, formalin-fixed and paraffin-embedded; qRT-PCR, quantitative real-time polymerase chain reaction; mPFS, median progression-free survival; mOS, median overall survival; mFFS, median failure-free survival; TTT, time to first treatment from diagnosis; BCR, B-cell receptor; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B cells; NA, not available; NR, not reached; nnMCL, non-nodal MCL; cMCL, classical MCL.
Initially, the proliferative gene signature was investigated in MCL as a quantitative integrator of multiple oncogenic aberrations (49), resulting in a better outcome predictor than single factor models based on individual oncogenic events. Four subgroups with significantly different median OS (6.7 vs. 3.3 vs. 2.3 vs. 0.8 years) were categorized. Nonetheless, as GEP requires fresh tissue, it is not applicable to everyday diagnostic routine, and Ki-67 staining was introduced as a surrogate index of cell proliferation, as it is easier to apply in clinical practice (14,15).
More recently, activation of B-cell receptor (BCR) and canonical NF-kB signaling was specifically described in MCL. The quantification of the BCR signaling strength was reflected by the expression of BCR-regulated genes and closely correlated with tumor proliferation. In particular, the autonomous signaling correlated with mutations and polymorphisms in these pathways and, thus, is apparently independent of microenvironment support. Activation of BCR identified a subset of patients with inferior survival after cytotoxic therapy. After a median follow-up of 7.5 years, the OS in patients with BCRhigh scores was 68%, compared to 96% for patients with BCRlow scores (HR =6.88, P=0.05) (7).
To provide an easy-to-use prognostic tool for the identification of high-risk MCL patients, Bomben et al. proposed a six-gene BCR signature to be assessed by qPCR (45). The analysis was performed in the context of the frontline FIL “MCL0208” phase III trial on CD19-selected peripheral blood (PB) cells and formalin-fixed paraffin-embedded (FFPE) samples from lymph node biopsies. Among this younger population, the signature targeting AKT3, BCL2, BTK, CD79B, PIK3CD, and SYK was able to identify a BCRhigh group characterized by discouraging outcomes, with a median PFS of 42 months versus “not reached” in BCRlow patients (P<0.01). Combining the BCR signature with the Ki-67 index achieved further refinement of outcome discrimination; the median PFS and OS were 21 and 47 months for BCRhigh with Ki-67 ≥30%, respectively, versus “not reached” for all other combinations (P<0.01 for PFS and P<0.05 for OS, respectively).
Recently, with technical improvements due to the availability of NanoString analysis on a digital platform, a new molecular assay to test the proliferation signature in FFPE samples was introduced in MCL (8). Scott et al. trained the assay using 47 FFPE biopsies and microarray gene expression data from matched fresh frozen biopsies as a gold standard. Subsequently, a model was developed using the expression of 17 proliferation genes to replicate the proliferation signature score described by Rosenwald et al. (49). This locked assay (“MCL35”) was validated in an independent cohort of 110 MCL patients. This signature defined groups of patients with significantly different OS, independent from MIPI score and treatment received (R-CHOP ± ASCT). Accordingly, patients were classified into three risk groups (high 26%, standard 29%, and low 45%) with significantly different median OS: 1.1, 2.6, and 8.6 years, respectively (P<0.001). Notably, the MCL35 assay clustered MCL cases with adverse “classical” biological characteristics, such as blastoid morphology and elevated Ki-67, as high-risk patients according to their proliferative signature (8). Finally, the strong prognostic value of this assay was validated in the prospective cohorts of the European MCL Network “MCL Younger” and “MCL Elderly” phase III clinical trials (46), as well as the Nordic “MCL2” and “MCL3” phase II trials (47). Alone or combined with MIPI or MIPI-c scores, the MCL35 assay could identify patients with dismal outcome despite intensified treatment.
In addition, a novel NanoString-based prognostic tool was recently described. The 16-gene signature “L-MCL16” is able to distinguish conventional (c) and leukemic non-nodal (nn) MCL (48); this clinical variant, characterized by splenomegaly, lymphocytosis, and no or minimal nodal involvement, has an overall excellent prognosis, even without treatment. Early and precise identification of these MCL patients would allow an alternative approach, rather than conventional, chemotherapy-based regimens. The L-MCL16 assay was applied to a cohort of 70 MCL patients with leukemic presentation, assigning 37% of cases to nnMCL and 56% to cMCL. These patient groups differed in some clinical and biological characteristics (i.e., nodal presentation, immunoglobulin heavy chain gene mutational status, genomic complexity), with nnMCL presenting significantly better survival than cMCL (3-year OS 92% vs. 69%; P<0.01). In summary, even though its application may be challenging in patients with low levels of leukemic disease, the L-MCL16 assay combined with clinical and genetic data will probably help better identify this peculiar, indolent entity, allowing patients to be spared unnecessary treatment.
Micro-RNA based prognostic tools
Another field of prognostication in MCL focuses on microRNA (miR) analysis. MiRs are short sequences of non-coding RNA implicated in the regulation of the expression of several genes responsible for different cellular functions and, namely, oncogenesis. Comparison of MCL samples to their normal counterparts (naive B cells) has identified differentially expressed miR with roles in cellular growth and survival pathways (50-53) in the last few years, and the clinical impact of miR in MCL has been studied in some small retrospective series; each study identified different miRs as being associated with outcome (Table 3) (50,55).
Table 3
| Patient series | Tissue (FFPE) | Involved miR | Method | Postulated function | Median OS |
|---|---|---|---|---|---|
| (54) | 50 LNs | miR-17-5p, miR-20a | RT-qPCR | Survival and apoptosis | The association between miR and high MYC expression identifies a poor survival group |
| (50) | 29 LNs, 1 spleen sample | miR-29 family | Microarray, RT-qPCR | Cell cycle control | Based on miR 29 family expression: low mOS 1.5 years; high mOS NR |
| (51) | 54 LNs; 82 LNs | miR-17-92 cluster | RT-qPCR, microarray (mRNA) | Chemoresistance and anti-apoptotic activity via PI3K/AKT pathway | Based on level of C13orf25: high mOS 1.06 years; low mOS 2.75 years |
| (52) | 30 LNs | 6-miRNA signature (high expression of miR129-3p, miR-135a, miR-146a, miR-424, and miR-450-5p and low expression of miR-222) | RT-qPCR array | Proliferative and microenvironment signature | Good risk group mOS 4 years; poor risk group mOS 2 years (P<0.05) |
| (55) | 23 LNs; 54 LNs | miR-20b | Microarray, RT-qPCR | Survival and proliferation | Based on expression level of miR20b: low mOS 5 years; high mOS 2.5 years (P=0.032) |
| (56) | 119 FFPE | miR-127-3p; miR-615-3p | TaqMan low-density arrays | NS | Ki67 and miR expression combined in one model: good mOS 46.3 months; intermediate mOS 18.8 months; poor mOS 9.5 months |
| (57) | 53 LNs; 12 tonsils; 2 colon; 1 stomach; 1 orbit; 1 parotid | miR-17-92 | RT-qPCR | Cell cycle control and apoptosis | 2 prognostic clusters: high SOX11/SOX12/miR19b/miR92a mOS 2 years; high SOX4/miR17/miR18a mOS NR (P<0.001) |
| (58) | 21 PBMCs (cd19+) | miR-223 | RT-qPCR | Cell proliferation and apoptosis | High expression mOS 36 months; low expression mOS 12 months (P=0.021) |
| (59) | 172 FFPE | miR-18b | miRNA assay and qRT-PCR | Proliferation and apoptosis | MIPI-miR-18b combined in one model for 3 risk classes: low mOS NR; intermediate mOS 7 years; high mOS 2 years (P=0.001) |
| (60) | 61 FFPE | miR-18b | qRT-PCR | Proliferation and apoptosis | MIPI-miR-18b combined in one model for 3 risk classes: low mOS NR; intermediate mOS 8.3 years; high mOS 1.6 years (P=0.000) |
FFPE, formalin-fixed paraffin embedded; LNs, lymph nodes; miR, micro-RNA; PBMC, peripheral blood mononuclear cell; RT-qPCR, quantitative reverse-transcribed polymerase chain reaction; GEP, gene expression profiling; NR, not reached; NS, not specified; mOS, median overall survival.
A distinctive miR signature was identified and validated in two small, retrospective cohorts of patients with aggressive lymphoma. Of this signature, two miRs (miR 127-3p and miR 615-3p) were significantly associated with OS in a training set of 119 MCL patients and validated in an independent cohort of 114 MCL patients. Moreover, the combined use of miR and classical prognostic factors (Ki-67 and MIPI) seemed to better identify high-risk patients (56-58).
Finally, Husby et al. investigated and validated the clinical effect of miR expression in two large prospective homogenously treated cohorts (59). In this study, 74 diagnostic MCL samples from the Nordic MCL2 trial were profiled for miRs, and prognostic miRs were validated in an independent series of 94 patients from the MCL3 trial. MiR-18b overexpression was able to identify patients with poor prognosis, and a new biological prognostic index was proposed combining miR-18b levels with MIPI-b (MIPI-b-miR), which identified high-risk patients in terms of both PFS and OS. These data were confirmed in the MCL2 population after 15 years of follow-up (60). Finally, the authors suggested that miR-18b may contribute to chemoresistance by decelerating cell proliferation.
Epigenomics and DNA methylation signatures
The study of epigenetic patterns using unbiased genome-wide approaches is reshaping our perception of the role of DNA methylation in cancer. The epigenetic landscape is assumed to play an increasingly important role in MCL, as increasing knowledge of the genetic basis of this lymphoma has not been able to explain the variability in its clinical course (61,62).
A systematic study of methyloma in 82 MCL patients revealed two major subtypes with distinct clinicobiological features (63). Patients characterized by a DNA methylation pattern more similar to germinal center-inexperienced B cells (i.e., hypomethylation of enhancers and transcribed regions) had significantly worse OS than the antigen-experienced group. The authors also found that the number of DNA methylation changes had a significant linear association with the clinical outcome, approximately doubling the risk of death with each 10,000 methylation changes. These data suggest that patients with more epigenetic changes have a worse clinical outcome that correlates with the acquisition of genetic changes and increased cell proliferation, particularly in cases transforming from leukemic, non-nodal, indolent MCL. However, more clinically oriented studies with a better characterized and homogeneously treated series are required to validate these findings before introducing epigenetic-based tools into the current risk stratification models of MCL.
How can we tailor therapy based on these biomarkers?
Despite many publications and validations of the prognostic impact of the different MIPI scores (Table 4) (1-3,64-68), none have yet been investigated as a treatment-tailoring tool in MCL, and therapeutic choices are still selected on the basis of age and comorbidities (69). Even though some new clinical entities with less aggressive behavior (i.e., MALT-like MCL) have been proposed recently (70), a de-escalation of treatment intensity for low-risk MIPI patients is still considered potentially harmful. Moreover, neither the blastoid morphology nor the Ki-67 index are used to drive different therapeutic choices, despite their strong association with poor outcomes (16). The only clinical trial specifically offering upfront single-agent high-dose cytarabine and rituximab for high-risk MIPI-b MCL rapidly stopped enrollment due to inefficacy (71).
Table 4
| Authors | MIPI | Risk factors | Clinical impact (mOS) |
|---|---|---|---|
| GLSG1996, GLSG2000, European MCL Trial1 (1) | MIPI | [0.03535 × age (years)] × age (years) + 0.6978 (if ECOG >1) + 1.367 × log10(LDH/ULN) + 0.9393 × log10(WBC count) | Low NR; intermediate 51 months; high 29 months |
| MCL Younger, MCL Elderly (2) | MIPI-s (simplified MIPI) | 0–3 points for each factor; Age (years); ECOG PS; LDH/ULN; WBC (109/L) | Low NR; intermediate NR; high 2.3 years (P<0.001) |
| GLSG1996, GLSG2000, European MCL Trial1 (1) | MIPI-b (biological MIPI) | MIPI score + 0.02142 × Ki-67 (%) | Low NR; intermediate 58 months; high 37 months |
| MCL Younger, MCL Elderly (3) | MIPI-c (combined MIPI) | MIPI risk classes divided by dichotomous (cut-off 30%) Ki67 | Low NR; low-Intermediate NR; high-Intermediate 5.1 years high 1 year (P<0.001) |
| MCL0208 FIL trial (21) | MIPI-g (genetic MIPI) | MIPI-c score + KMT2D/TP53 disruption | Low 4-year OS 94%; intermediate 4-year OS 65%; high 4-year OS 45% |
| MCL2/MCL3 trials (60) | MIPI-b-miR (miRNA-18b MIPI) | MIPI-b score + 0.58317 × log-fold-change of miR-18b | Low NR; intermediate 8.3 years; high 1.6 years (P<0.001) |
MCL, mantle cell lymphoma; MIPI, MCL International Prognostic Index; mOS, median overall survival; NR, not reached; GLSG, German Low Grade Lymphoma Study Group; FIL, Fondazione Italiana Linfomi; miR, micro-RNA; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; WBC, white blood cells; KMT2D, lysine methyltransferase 2D.
Given the favorable outcome of MRD clearance in MCL (5,22), and considering the proposed MRD-driven pre-emptive strategies (72,73), some ongoing clinical trials (NCT02896582 and NCT03267433) are investigating modulation of the maintenance therapy based on MRD results (74). Some preliminary data suggest that maintenance therapy may benefit MRD-negative patients as well, meaning that the preservation of MRD-negativity rather than the conversion of MRD-positivity may be a valuable goal of current post-induction therapies (10). As no clear survival benefit of MRD-driven strategies has been demonstrated thus far, more data are required before the modulation of maintenance treatments based on MRD results (69,75). Morever new techniques for MRD detection are emerging. Next-generation Sequencing (NGS) techniques (76) and cell free circulating DNA (cfDNA) monitoring (77,78) are able to overcome some limitations of standard RQ-PCR, but their application is not yet validated or standardized.
In addition, increasing evidence from mutational studies is challenging the current scenario of MCL and may soon change the clinical management of these patients. The prognostic value of TP53 disruption (both mutations and deletions) has been uniformly confirmed by independent research groups (19,21,28). TP53-disrupted MCL presents dismal outcomes independent from classical prognosticators (e.g., MIPI, clinical response, or MRD negativity) and seems to not benefit from the addition of lenalidomide (29,79). Unfortunately, data on the impact of BTK inhibitors in these patients are scarce. In one study, despite an overall response in 11 out of 20 (55%) relapsing patients treated with ibrutinib monotherapy, responses to this therapy were short-lasting (median PFS 4 months) (80). However, some promising data come from combinations with venetoclax or with rituximab and lenalidomide [up to 50–64% with complete response (CR) and no difference in PFS compared to WT], but the series of TP53-disrupted patients described in these phase II trials (n=12 and 11, respectively) and the median follow-up (16 and 18 months, respectively) are too limited to draw firm conclusions (81,82).
Therefore, although prognostic, TP53 disruption has not yet been established as a predictive biomarker sufficient to drive novel therapeutic approaches. Nevertheless, it is becoming increasingly clear that MCL patients with TP53 disruption should be included in front-line clinical trials exploring novel targeted drugs rather than receiving standard immunochemotherapy. In this regard, the EuMCLNet “TRIANGLE” phase III trial is investigating the integration of ibrutinib in first-line high-dose immunochemotherapy in younger patients (EudraCT 2014-001363-12), but no mutational stratification is being applied. Other BTK inhibitors are currently being evaluated, such as acalabrutinib in association with bendamustine-rituximab in a phase II trial (NCT03863184), and zanubrutinib vs. bendamustine-rituximab in a phase III trial (NCT04002297). Interestingly, the recruiting FIL “V-RBAC” phase II trial (EudraCT No. 2017-004628-31) is offering consolidation with venetoclax after four cycles of R-BAC500 (17) to elderly patients characterized by TP53 disruption, Ki-67 ≥30%, or blastoid variant.
Finally, although the real effectiveness in these high-risk patients requires investigation, front-line consolidation with reduced-intensity conditioning allogeneic transplantation may be considered in younger and fit patients (83). A similar role may be claimed by CAR-T consolidation as soon as this novel approach is available (84).
Therapeutic decisions on the basis of genomic aberrations other than TP53 disruption should be discouraged. KMT2D mutations are the only other genomic biomarker with an independent, negative impact on OS that has been validated externally, though in a small series (21); therefore, the authors proposed the new MIPI-g (integrating both TP53 disruption and KMT2D mutations) as a useful tool for selecting high-risk MCL patients for future, “tailored” experimental strategies. However, the diagnostic test for KMT2D mutations is currently not standardized and not available in clinical practice. Moreover, no data are available on the impact of new drugs on KMT2D mutations. Finally, even if both CDKN2 deletions and NOTCH1 mutations have been described as detrimental in terms of survival, their prognostic impact may not be independent, as they are often associated with TP53 disruptions (19,21,28). Finally, other mutations have been proposed as predictive markers for targeted approaches, but larger series confirmation is missing (85,86). For example, a functional deficit of CDKN2 seems to attenuate the efficacy of the new CD4/6 inhibitor palbociclib, which was tested in a phase II US trial (NCT03478514) (87).
Regarding immunohistochemistry markers, although both SOX11 and p53 staining is recommended in clinical routine (37), neither should be used for tailoring treatment. Diagnosis of leukemic nnMCL is made independent of SOX11 status, and SOX11-negativity has no relevant clinical significance in classical MCL (9). However, although the clinical value of p53 expression has been validated, its use is not yet widespread, and strict assessment guidelines (38) need to be followed.
Finally, the recently developed GEP-based assays represent easily applicable and highly promising treatment-tailoring tools, particularly when available on the widespread NanoString platform (7,8,45,48). The analytical and clinical validity of the MCL35 (8) and L-MCL16 (48) assays prompts indicates they are reliable biomarkers for risk-adapted clinical trials. L-MCL16 is able to reliably distinguish indolent MCL from classical subtypes, even if the tool is mainly conceived for leukemic cases, as the analysis is done on PB, whereas MCL35 is conceived for highly infiltrated FFPE samples. However, roughly one-third of the non-nodal, indolent cases carry a high number of CNAs and a similar prognosis as classical MCL patients. Therefore, it is not untimely to foresee a tailored therapeutic approach in which patients with high CNA, regardless of subtype, are prioritized in trials (or treatments) with novel agents, whereas those with classical MCL and low CNA receive standard immunochemotherapy (88). On the other hand, patients with leukemic nnMCL and low CNA may be either observed or enrolled into innovative clinical trials in which treatment mechanisms are evaluated over time (e.g., “master protocols”) (89).
Interestingly, a couple of GEP studies focusing on BCR-related signatures have shown that patients over-expressing such genes have worse prognosis, suggesting that they might be ideal candidates to receive first-line treatment with BTK inhibitors (7,8,45). Nevertheless, to test this exciting hypothesis, these signatures should first be investigated in prospective clinical trials with BTK inhibitors (e.g., the TRIANGLE trial).
In summary, some words of caution have to be added concerning these highly promising GEP-based tools before considering them for treatment-tailoring. Overall, a general limitation of gene expression signatures is that their prognostic significance is highly dependent on the specific treatment received, as demonstrated in follicular lymphoma (44,90). Therefore, the concept of current GEP tools should be limited to MCL patients receiving conventional immunochemotherapy with R-CHOP or high-dose cytarabine-containing schedules (± ASCT); thus, any putative impact on bendamustine combinations or new drugs still needs to be demonstrated (46,47). Moreover, some technical limitations need to be overcome before the effective introduction of these biomarkers into clinical practice: low infiltration (<60%) FFPE samples and bone marrow samples are currently not suitable for MCL35 or L-MCL16. Moreover, interlaboratory standardization is still needed for these tools, though feasible on NanoString technology. Until these issues are covered, GEP-based tools still remain limited to the context of translational research.
Conclusions
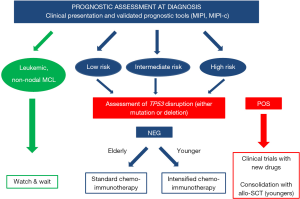
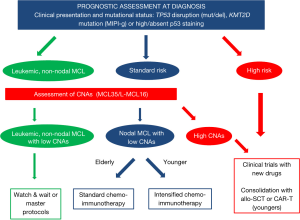
In conclusion, this review discussed the current scenario of prognostic tools in MCL and their possible application in tailoring treatment in the context of both clinical trials and, more importantly, real life. Although many promising biomarkers were established during the last 10 years (Figure 1), the authors’ main aim was to focus on the few prognostic tools, such as TP53 disruption, that clinicians can start to use right now in the daily management of MCL patients (Figure 2). Moreover, a schematic picture of the most promising new biomarkers that may soon gain clinical use is presented in Figure 3.
In 2020, MCL is still a challenge for both clinical and translational hematologists, given its rarity and its heterogeneous natural course. However, thanks to the continuous contribution of novel biological insights, and the international collaboration in conducting innovative clinical trials, both academic and industry driven, a real opportunity to pursue personalized medicine in clinical practice is being presented, and should be the main goal of MCL research in the present decade.
Acknowledgments
Funding: This work was supported by Fondi di Ricerca Locale, Università degli Studi di Torino, Italy; Fondazione Neoplasie Del Sangue (Fo.Ne.Sa), Torino, Italy; Fondazione CRT (project codes: 2016.0677 and 2018.1284), Torino, Italy; and the Gilead Fellowship Program 2019, Milano, Italy.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Lymphoma for the series “Future Directions for Mantle Cell Lymphoma”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-2018-mcl-010). The series “Future Directions for Mantle Cell Lymphoma” was commissioned by the editorial office without any funding or sponsorship. MD served as the unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hoster E, Dreyling M, Klapper W, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood 2008;111:558-65. [Crossref] [PubMed]
- Hoster E, Klapper W, Hermine O, et al. Confirmation of the Mantle-Cell Lymphoma International Prognostic Index in Randomized Trials of the European Mantle-Cell Lymphoma Network. J Clin Oncol 2014;32:1338-46. [Crossref] [PubMed]
- Hoster E, Rosenwald A, Berger F, et al. Prognostic Value of Ki-67 Index, Cytology, and Growth Pattern in Mantle-Cell Lymphoma: Results From Randomized Trials of the European Mantle Cell Lymphoma Network. J Clin Oncol 2016;34:1386-94. [Crossref] [PubMed]
- Pott C, Schrader C, Gesk S, et al. Quantitative assessment of molecular remission after high-dose therapy with autologous stem cell transplantation predicts long-term remission in mantle cell lymphoma. Blood 2006;107:2271-8. [Crossref] [PubMed]
- Pott C, Hoster E, Delfau-Larue MH, et al. Molecular remission is an independent predictor of clinical outcome in patients with mantle cell lymphoma after combined immunochemotherapy: a European MCL intergroup study. Blood 2010;115:3215-23. [Crossref] [PubMed]
- Beà S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A 2013;110:18250-5. [Crossref] [PubMed]
- Saba NS, Liu D, Herman SEM, et al. Pathogenic role of B-cell receptor signaling and canonical NF-κB activation in mantle cell lymphoma. Blood 2016;128:82-92. [Crossref] [PubMed]
- Scott DW, Abrisqueta P, Wright GW, et al. New Molecular Assay for the Proliferation Signature in Mantle Cell Lymphoma Applicable to Formalin-Fixed Paraffin-Embedded Biopsies. J Clin Oncol 2017;35:1668-77. [Crossref] [PubMed]
- Aukema SM, Hoster E, Rosenwald A, et al. Expression of TP53 is associated with the outcome of MCL independent of MIPI and Ki-67 in trials of the European MCL Network. Blood 2018;131:417-20. [Crossref] [PubMed]
- Callanan MB, Delfau MH, Macintyre E, et al. Predictive Power of Early, Sequential MRD Monitoring in Peripheral Blood and Bone Marrow in Patients with Mantle Cell Lymphoma Following Autologous Stem Cell Transplantation with or without Rituximab Maintenance; Interim Results from the LyMa-MRD Project, Conducted on Behalf of the Lysa Group. Blood 2015;126:338. [Crossref]
- Ferrero S, Daniela B, Lo Schirico M, et al. Comprehensive Minimal Residual Disease (MRD) Analysis of the Fondazione Italiana Linfomi (FIL) MCL0208 Clinical Trial for Younger Patients with Mantle Cell Lymphoma: A Kinetic Model Ensures a More Refined Risk Stratification. Blood 2018;132:920. [Crossref]
- Hermine O, Hoster E, Walewski J, et al. Addition of high-dose cytarabine to immunochemotherapy before autologous stem-cell transplantation in patients aged 65 years or younger with mantle cell lymphoma (MCL Younger): a randomised, open-label, phase 3 trial of the European Mantle Cell Lymphoma Network. Lancet 2016;388:565-75. [Crossref] [PubMed]
- Kluin-Nelemans HC, Hoster E, Hermine O, et al. Treatment of Older Patients with Mantle-Cell Lymphoma. N Engl J Med 2012;367:520-31. [Crossref] [PubMed]
- Tiemann M, Schrader C, Klapper W, et al. Histopathology, cell proliferation indices and clinical outcome in 304 patients with mantle cell lymphoma (MCL): a clinicopathological study from the European MCL Network. Br J Haematol 2005;131:29-38. [Crossref] [PubMed]
- Determann O, Hoster E, Ott G, et al. Ki-67 predicts outcome in advanced-stage mantle cell lymphoma patients treated with anti-CD20 immunochemotherapy: results from randomized trials of the European MCL Network and the German Low Grade Lymphoma Study Group. Blood 2008;111:2385-7. [Crossref] [PubMed]
- Dreyling M, Ferrero S, Vogt N, et al. New Paradigms in Mantle Cell Lymphoma: Is It Time to Risk-Stratify Treatment Based on the Proliferative Signature? Clin Cancer Res 2014;20:5194-206. [Crossref] [PubMed]
- Visco C, Chiappella A, Nassi L, et al. Rituximab, bendamustine, and low-dose cytarabine as induction therapy in elderly patients with mantle cell lymphoma: a multicentre, phase 2 trial from Fondazione Italiana Linfomi. Lancet Haematol 2017;4:e15-23. [Crossref] [PubMed]
- Dreyling M, Klapper W, Rule S. Blastoid and pleomorphic mantle cell lymphoma: still a diagnostic and therapeutic challenge! Blood 2018;132:2722-9. [Crossref] [PubMed]
- Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood 2017;130:1903-10. [Crossref] [PubMed]
- Derby L, Pendurti G, Deeb G, et al. Blastoid Variant of Mantle Cell Lymphoma (MCL) Is Associated with P53 Abnormalities and Have a Shorter Progression-Free Survival (PFS) and Overall Survival (OS) Despite Upfront Chemo-Immunotherapy Followed by High Dose Chemotherapy and Autologous Stem Cell Support (HDC-ASCS). Blood 2010;116:1773. [Crossref]
- Ferrero S, Rossi D, Rinaldi A, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica 2020;105:1604-12. [Crossref] [PubMed]
- Kolstad A, Pedersen LB, Eskelund CW, et al. Molecular Monitoring after Autologous Stem Cell Transplantation and Preemptive Rituximab Treatment of Molecular Relapse; Results from the Nordic Mantle Cell Lymphoma Studies (MCL2 and MCL3) with Median Follow-Up of 8.5 Years. Biol Blood Marrow Transplant 2017;23:428-35. [Crossref] [PubMed]
- Halldórsdóttir AM, Lundin A, Murray F, et al. Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia 2011;25:1904-8. [Crossref] [PubMed]
- Xu-Monette ZY, Wu L, Visco C, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2012;120:3986-96. [Crossref] [PubMed]
- Nordström L, Sernbo S, Eden P, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma - a Nordic Lymphoma Group study. Br J Haematol 2014;166:98-108. [Crossref] [PubMed]
- Rossi D, Rasi S, Fabbri G, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012;119:521-9. [Crossref] [PubMed]
- Wang L, Tang G, Medeiros LJ, et al. MYC rearrangement but not extra MYC copies is an independent prognostic factor in patients with mantle cell lymphoma. Haematologica 2020; Epub ahead of print. [Crossref] [PubMed]
- Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood 2015;126:604-11. [Crossref] [PubMed]
- Eskelund CW, Albertsson-Lindblad A, Kolstad A, et al. Lenalidomide plus bendamustine-rituximab does not overcome the adverse impact of TP53 mutations in mantle cell lymphoma. Haematologica 2018;103:e541-3. [Crossref] [PubMed]
- Zhang Y, Xiong Y, Yarbrough WG. ARF Promotes MDM2 Degradation and Stabilizes p53: ARF-INK4a Locus Deletion Impairs Both the Rb and p53 Tumor Suppression Pathways. Cell 1998;92:725-34. [Crossref] [PubMed]
- Chen YH, Gao J, Fan G, Peterson LC. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol 2010;23:105-12. [Crossref] [PubMed]
- Vegliante MC, Palomero J, Pérez-Galán P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood 2013;121:2175-85. [Crossref] [PubMed]
- Fernàndez V, Salamero O, Espinet B, et al. Genomic and Gene Expression Profiling Defines Indolent Forms of Mantle Cell Lymphoma. Cancer Res 2010;70:1408-18. [Crossref] [PubMed]
- Navarro A, Clot G, Royo C, et al. Molecular Subsets of Mantle Cell Lymphoma Defined by the IGHV Mutational Status and SOX11 Expression Have Distinct Biologic and Clinical Features. Cancer Res 2012;72:5307-16. [Crossref] [PubMed]
- Nygren L, Baumgartner Wennerholm S, Klimkowska M, et al. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood 2012;119:4215-23. [Crossref] [PubMed]
- Condoluci A, Rossi D, Zucca E, et al. Toward a Risk-Tailored Therapeutic Policy in Mantle Cell Lymphoma. Curr Oncol Rep 2018;20:79. [Crossref] [PubMed]
- Dreyling M, Hoster E, Unterhalt M, et al. Clinical Outcome of Mantle Cell Lymphoma Patients with High Risk Biology (high Ki-67, blastic MCL, or high p53 expression). Blood 2019;134:3996. [Crossref]
- Croci GA, Hoster E, Beà S, et al. Reproducibility of histologic prognostic parameters for mantle cell lymphoma: cytology, Ki67, p53 and SOX11. Virchows Arch 2020;477:259-67. [Crossref] [PubMed]
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503-11. [Crossref] [PubMed]
- Rosenwald A, Wright G, Chan WC, et al. The Use of Molecular Profiling to Predict Survival after Chemotherapy for Diffuse Large-B-Cell Lymphoma. N Engl J Med 2002;346:1937-47. [Crossref] [PubMed]
- Lenz G, Wright G, Dave SS, et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. N Engl J Med 2008;359:2313-23. [Crossref] [PubMed]
- Ciavarella S, Vegliante MC, Fabbri M, et al. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann Oncol 2018;29:2363-70. [Crossref] [PubMed]
- Dave SS, Wright G, Tan B, et al. Prediction of Survival in Follicular Lymphoma Based on Molecular Features of Tumor-Infiltrating Immune Cells. N Engl J Med 2004;351:2159-69. [Crossref] [PubMed]
- Huet S, Tesson B, Jais JP, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol 2018;19:549-61. [Crossref] [PubMed]
- Bomben R, Ferrero S, D’Agaro T, et al. A B-cell receptor-related gene signature predicts survival in mantle cell lymphoma: results from the Fondazione Italiana Linfomi MCL-0208 trial. Haematologica 2018;103:849-56. [Crossref] [PubMed]
- Rauert-Wunderlich H, Mottok A, Scott DW, et al. Validation of the MCL 35 gene expression proliferation assay in randomized trials of the European Mantle Cell Lymphoma Network. Br J Haematol 2019;184:616-24. [Crossref] [PubMed]
- Holte H, Beiske K, Boyle M, et al. The MCL35 gene expression proliferation assay predicts high-risk MCL patients in a Norwegian cohort of younger patients given intensive first line therapy. Br J Haematol 2018;183:225-34. [Crossref] [PubMed]
- Clot G, Jares P, Giné E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood 2018;132:413-22. [Crossref] [PubMed]
- Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 2003;3:185-97. [Crossref] [PubMed]
- Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010;115:2630-9. [Crossref] [PubMed]
- Rao E, Jiang C, Ji M, et al. The miRNA-17-92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia 2012;26:1064-72. [Crossref] [PubMed]
- Iqbal J, Shen Y, Liu Y, et al. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood 2012;119:4939-48. [Crossref] [PubMed]
- Husby S, Geisler C, Grønbæk K. MicroRNAs in mantle cell lymphoma. Leuk Lymphoma 2013;54:1867-75. [Crossref] [PubMed]
- Navarro A, Bea S, Fernandez V, et al. MicroRNA Expression, Chromosomal Alterations, and Immunoglobulin Variable Heavy Chain Hypermutations in Mantle Cell Lymphomas. Cancer Res 2009;69:7071-8. [Crossref] [PubMed]
- Di Lisio L, Gómez-López G, Sánchez-Beato M, et al. Mantle cell lymphoma: transcriptional regulation by microRNAs. Leukemia 2010;24:1335-42. [Crossref] [PubMed]
- Goswami RS, Atenafu EG, Xuan Y, et al. MicroRNA signature obtained from the comparison of aggressive with indolent non-Hodgkin lymphomas: potential prognostic value in mantle-cell lymphoma. J Clin Oncol 2013;31:2903-11. [Crossref] [PubMed]
- Roisman A, Huamán Garaicoa F, Metrebian F, et al. SOXC and MiR17-92 gene expression profiling defines two subgroups with different clinical outcome in mantle cell lymphoma: SOXC and M I R17-92 Gene Expression. Genes Chromosomes Cancer 2016;55:531-40. [Crossref] [PubMed]
- Zhou K, Feng X, Wang Y, et al. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp Hematol 2018;58:27-34.e1. [Crossref] [PubMed]
- Husby S, Ralfkiaer U, Garde C, et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood 2015;125:2669-77. [Crossref] [PubMed]
- Eskelund CW, Kolstad A, Jerkeman M, et al. 15-year follow-up of the Second Nordic Mantle Cell Lymphoma trial (MCL2): prolonged remissions without survival plateau. Br J Haematol 2016;175:410-8. [Crossref] [PubMed]
- Enjuanes A, Albero R, Clot G, et al. Genome-wide methylation analyses identify a subset of mantle cell lymphoma with a high number of methylated CpGs and aggressive clinicopathological features. Int J Cancer 2013;133:2852-63. [Crossref] [PubMed]
- Halldórsdóttir AM, Kanduri M, Marincevic M, et al. Mantle cell lymphoma displays a homogenous methylation profile: A comparative analysis with chronic lymphocytic leukemia. Am J Hematol 2012;87:361-7. [Crossref] [PubMed]
- Queirós AC, Beekman R, Vilarrasa-Blasi R, et al. Decoding the DNA Methylome of Mantle Cell Lymphoma in the Light of the Entire B Cell Lineage. Cancer Cell 2016;30:806-21. [Crossref] [PubMed]
- Geisler CH, Kolstad A, Laurell A, et al. The Mantle Cell Lymphoma International Prognostic Index (MIPI) is superior to the International Prognostic Index (IPI) in predicting survival following intensive first-line immunochemotherapy and autologous stem cell transplantation (ASCT). Blood 2010;115:1530-3. [Crossref] [PubMed]
- Smith SD, Hsi ED, Bolwell BJ, et al. Validation of the Mantle Cell Lymphoma Prognostic Index (MIPI): A Valuable Tool for Risk Stratification in Mantle Cell Lymphoma. Blood 2009;114:2703. [Crossref]
- Chiappella A, Botto B, Marmont F, et al. Validation of Mantle Cell International Prognostic Index (MIPI) in Mantle Cell Lymphoma (MCL) in a Retrospective Single Institution Series. Blood 2008;112:2828. [Crossref]
- Salek D, Vesela P, Boudova L, et al. Retrospective analysis of 235 unselected patients with mantle cell lymphoma confirms prognostic relevance of Mantle Cell Lymphoma International Prognostic Index and Ki-67 in the era of rituximab: long-term data from the Czech Lymphoma Project Database. Leuk Lymphoma 2014;55:802-10. [Crossref] [PubMed]
- van de Schans SAM, Janssen-Heijnen MLG, et al. Validation, revision and extension of the Mantle Cell Lymphoma International Prognostic Index in a population-based setting. Haematologica 2010;95:1503-9. [Crossref] [PubMed]
- Dreyling M, Campo E, Hermine O, et al. Newly diagnosed and relapsed mantle cell lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv62-71. [Crossref] [PubMed]
- Morello L, Rattotti S, Giordano L, et al. Mantle Cell Lymphoma of Mucosa-Associated Lymphoid Tissue: A European Mantle Cell Lymphoma Network Study. Hemasphere 2019;4:e302 [Crossref] [PubMed]
- Laurell A, Kolstad A, Jerkeman M, et al. High dose cytarabine with rituximab is not enough in first-line treatment of mantle cell lymphoma with high proliferation: early closure of the Nordic Lymphoma Group Mantle Cell Lymphoma 5 trial. Leuk Lymphoma 2014;55:1206-8. [Crossref] [PubMed]
- Andersen NS, Pedersen LB, Laurell A, et al. Pre-emptive treatment with rituximab of molecular relapse after autologous stem cell transplantation in mantle cell lymphoma. J Clin Oncol 2009;27:4365-70. [Crossref] [PubMed]
- Ferrero S, Monitillo L, Mantoan B, et al. Rituximab-based pre-emptive treatment of molecular relapse in follicular and mantle cell lymphoma. Ann Hematol 2013;92:1503-11. [Crossref] [PubMed]
- Dreyling M, Ferrero SEuropean Mantle Cell Lymphoma Network. The role of targeted treatment in mantle cell lymphoma: is transplant dead or alive?. Haematologica 2016;101:104-14. [Crossref] [PubMed]
- Ferrero S, Dreyling MEuropean Mantle Cell Lymphoma Network. Minimal residual disease in mantle cell lymphoma: are we ready for a personalized treatment approach? Haematologica 2017;102:1133-6. [Crossref] [PubMed]
- Ladetto M, Brüggemann M, Monitillo L, et al. Next-generation sequencing and real-time quantitative PCR for minimal residual disease detection in B-cell disorders. Leukemia 2014;28:1299-307. [Crossref] [PubMed]
- Roschewski M, Staudt LM, Wilson WH. Dynamic monitoring of circulating tumor DNA in non-Hodgkin lymphoma. Blood 2016;127:3127-32. [Crossref] [PubMed]
- Kurtz DM, Green MR, Bratman SV, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015;125:3679-87. [Crossref] [PubMed]
- Ferrero S, Zaccaria GM, Grimaldi D, et al. Focus on patients with TP53 disruption in the Fondazione Italiana Linfomi (FIL) MCL0208 trial: uniform poor outcome, regardless of baseline predictors, MRD status and lenalidomide maintenance. EHA2020 EP1160.
- Rule S, Dreyling M, Goy A, et al. Ibrutinib for the treatment of relapsed/refractory mantle cell lymphoma: extended 3.5-year follow up from a pooled analysis. Haematologica 2019;104:e211-4. [Crossref] [PubMed]
- Tam CS, Anderson MA, Pott C, et al. Ibrutinib plus Venetoclax for the Treatment of Mantle-Cell Lymphoma. N Engl J Med 2018;378:1211-23. [Crossref] [PubMed]
- Jerkeman M, Eskelund CW, Hutchings M, et al. Ibrutinib, lenalidomide, and rituximab in relapsed or refractory mantle cell lymphoma (PHILEMON): a multicentre, open-label, single-arm, phase 2 trial. Lancet Haematol 2018;5:e109-16. [Crossref] [PubMed]
- Dreger P, Michallet M, Bosman P, et al. Ibrutinib for bridging to allogeneic hematopoietic cell transplantation in patients with chronic lymphocytic leukemia or mantle cell lymphoma: a study by the EBMT Chronic Malignancies and Lymphoma Working Parties. Bone Marrow Transplant 2019;54:44-52. [Crossref] [PubMed]
- Wang M, Pruteanu I, Cohen AD, et al. Identification and Validation of Predictive Biomarkers to CD19- and BCMA-Specific CAR T-Cell Responses in CAR T-Cell Precursors. Blood 2019;134:622. [Crossref]
- Zhao X, Lwin T, Silva A, et al. Unification of de novo and acquired ibrutinib resistance in mantle cell lymphoma. Nat Commun 2017;8:14920. [Crossref] [PubMed]
- Agarwal R, Chan YC, Tam CS, et al. Dynamic molecular monitoring reveals that SWI–SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med 2019;25:119-29. [Crossref] [PubMed]
- Green JL, Okerberg ES, Sejd J, et al. Direct CDKN2 Modulation of CDK4 Alters Target Engagement of CDK4 Inhibitor Drugs. Mol Cancer Ther 2019;18:771-9. [Crossref] [PubMed]
- Martin P. A tale of two mantle cell lymphomas. Blood 2018;132:347-8. [Crossref] [PubMed]
- Biankin AV, Piantadosi S, Hollingsworth SJ. Patient-centric trials for therapeutic development in precision oncology. Nature 2015;526:361-70. [Crossref] [PubMed]
- Bolen C, Hiddemann W, Marcus R, et al. S100 Treatment-dependence of high-risk gene expression signatures in de novo follicular lymphoma. HemaSphere 2019;3: [Crossref]
Cite this article as: Ferrero S, Grimaldi D, Dreyling M; European Mantle Cell Lymphoma Network. Tailored treatment in mantle cell lymphoma. Ann Lymphoma 2020;4:12.


