
Genetic heterogeneity in follicular lymphoma
Introduction
Follicular lymphoma (FL) is driven by the accumulation of clonal neoplastic cells that closely mimic several phenotypic and functional attributes of their normal counterparts, germinal centre (GC) B-cells. There is a recognised heterogeneity in both clinical phenotype and disease outcome. On one end of the scale, there are patients with early-stage disease, half of which have long durable remissions with therapy, those that can be managed expectantly for many years and a rare minority with spontaneous remission. At the other end, there are patients who follow the prototypical relapsing-remitting disease course and additional high risk cohorts that progress rapidly within 2 years (POD24) (1,2) or histologically transform to high grade lymphoma, usually diffuse large B-cell lymphoma (DLBCL) (3). Whilst the introduction of anti-CD20 monoclonal antibodies, such as rituximab since the late 1990s, has significantly improved prognosis in FL, there is a recognition that high-risk patients who progress during or after therapy and those that undergo transformation require a different and perhaps more aggressive treatment strategy (2). Hypothetically, accurate identification of these high-risk subsets at diagnosis could enable risk-adapted therapeutic intervention, providing the most effective, targeted therapies to high-risk patients whilst also identifying those with low-risk clinical phenotypes might mean that therapy could be de-escalated to avoid over-treatment in a similar vein to the strategies employed in Hodgkin’s lymphoma. Current prognostic tools, based on clinical information (4,5) and those that incorporate mutations or gene expression data (6,7), can stratify patients into different risk groups but have not demonstrated a high enough prognostic accuracy to be routinely implemented in clinical practice (8,9).
This variability in clinical trajectories has perhaps hinted for decades at the underlying biological heterogeneity. The advent of next generation sequencing (NGS) has enabled more powerful elucidation of the breadth of accumulating genetic events that occur alongside the pathognomonic t(14;18) translocation as well as a better understanding of the extent of the genetic diversity, not only among FL from different patients (inter-tumour heterogeneity) but also within individual patient’s tumours (intra-tumour heterogeneity). This added insight at the intra-tumoral level permits the tracing of the lymphoma’s life history and patterns of clonal evolution.
This review will focus on the recent developments in our understanding of the mutational landscape and how it contributes to FL pathogenesis and the genetic complexities and tumour dynamics in FL uncovered by spatial and temporal genomic profiling with NGS-based technologies. We also discuss the potential and promise of these insights in terms of patient stratification and targeted therapeutic strategies.
Pre-NGS era: t(14;18) and broad genomic changes
The t(14;18) somatic translocation (10) is considered an early genetic event, occurring in approximately 85% of patients (11) and leads to BCL2 overexpression by bringing the anti-apoptotic BCL2 protein under the control of the immunoglobulin heavy chain (IGH) enhancer. In the majority of cases, these translocations occur at the major breakpoint region (mbr), although other breakpoint sites such as the minor cluster region (mcr) downstream of the mbr, the intermediate cluster region (icr), 5’mcr and 3’BCL2 are also observed at lower frequencies (12-17). Evidence over the years demonstrate that the t(14;18) translocation alone is insufficient for malignant transformation such as the 10–15% cohort of patients with t(14;18)-negative FL and the detection of low levels of t(14;18)-positivity reported in the peripheral blood of healthy individuals (18-20). Notably much elevated levels of circulating t(14;18)-positive B-cells may represent a reservoir of pre-malignant FL precursors as these individuals have a higher propensity of developing overt FL (21). Whilst the t(14;18) translocation is clearly a critical early event in conferring risk and initiating lymphomagenesis, further genetic events are required for the development of overt FL.
Genomic copy-number heterogeneity occurs extensively within FL tumours. These were initially identified by lower resolution approaches including cytogenetics, array CGH and DNA microarrays which led to the identification of recurrent copy number aberrations (CNAs) and copy neutral loss of heterozygosity (cnLOH) in FL. Commonly observed CNAs include gains in 1q, 2p, 7, 8, 12q, 18q, X and deletions of 1p36, 6q, 10q, 13p, 17p (22-26). Due to the large genomic regions encompassed by these chromosomal changes, it was not always possible to identify the precise target genes affected within these regions that contributed to FL pathogenesis. Some exceptions include the identification of TNFRSF14 within the frequently deleted 1p36 region (27), EPHA7 uncovered as one of the tumour suppressor genes within the commonly deleted 6q region (28) and the amplification of REL and MYC oncogenes within the 2p and 8q24 regions, respectively (22,24,25). The degree of chromosomal and CN heterogeneity varies between patients, inter-tumour heterogeneity, as well as between diagnostic, relapsed and transformed biopsies within the same patient, intra-tumour heterogeneity. Cytogenetic analysis of paired FL and transformed FL (tFL) showed increased complexity of genomic aberrations associated with transformation (22,24,25). Although some specific CNAs have been associated with inferior prognosis [such as 1p36 and 6q (23,25)] and/or increased risk of transformation [such as 3q27, 9p21, 11 and 15q (22)], there has been considerable variability and lack of reproducibility across studies due to the heterogeneous cohorts studied across different treatment eras. As CNAs in FL can disrupt hundreds of genes compared to single gene mutations, they likely contribute significantly to the genomic instability that acts as a fuel for tumour evolution and the many alternate evolutionary trajectories. However, CNAs represent only one of the factors that contribute to FL genomic heterogeneity, its initiation and tumour evolution.
Inter-tumour heterogeneity—mutational landscape of FL
NGS has been instrumental in the last decade in providing a finalised mutational catalogue of coding genes in FL. A number of biological pathways including epigenetic regulation and key signalling networks are dysregulated by the acquisition of recurrent gene mutations in FL. Figure 1 provides a summary of the main biological pathways affected in FL.
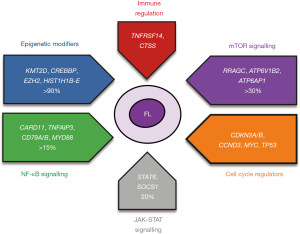
Mutations in epigenetic regulators—a defining hallmark of FL
Epigenetic regulatory mechanisms are commonly hijacked in tumorigenesis as they form an important part of a carefully choreographed gene regulation circuitry that permits the required sets of genes to be switched ‘on’ and ‘off’ within a particular cell at a specific time. A key functional unit of this circuitry, chromatin, a complex of DNA and histone proteins, exists in two main states: condensed transcriptionally repressed heterochromatin and transcriptionally active euchromatin. Epigenetic regulation between these two chromatin states occurs through a number of mechanisms including DNA methylation, histone post-translational modifications (PTMs) and chromatin remodelling.
A rather surprising finding from early NGS studies in FL was the high prevalence of mutations in epigenetic regulators, particularly those involved in histone PTMs. Approximately 90% of FLs carry one or more mutations in genes involved in epigenetic regulation through histone modifications (KMT2D, CREBBP and EZH2) and chromatin remodelling (HIST1H1B-E, ARID1A) (29).
KMT2D (also known as MLL2), a lysine-specific histone (H3K4) methyltransferase, is the most common histone-modifying enzyme mutated in FL, occurring in approximately 60–80% of patients. The mutations are typically inactivating in nature, leading to loss of the protein. Loss of KMT2D in conditional knock-out mouse models lead to a reduction in global H3K4 methylation, promoting a block at the GC stage of development and enhanced tumour suppressor gene expression (30,31).
Inactivating mutations in the histone acetyltransferase (HAT) enzymes CREBBP and, less commonly, EP300 (a structural paralog to CREBBP) occur in up to 70% and 15% of cases, respectively. The mutations affecting CREBBP are either missense (mostly clustered within the catalytic HAT domain), truncating or deleterious in nature. CREBBP and EP300 regulate gene expression by catalysing the acetylation of lysine residues in both histone and non-histone proteins. Transcriptionally, CREBBP-mutant mice and human tumours display focal depletion of H3K27 acetylation at gene enhancers central to GC development, B-cell receptor signalling and antigen presentation (29,32,33). Decreased MHC II expression resulting from loss-of-function CREBBP mutations limits antigen presentation by tumour B-cells with reduction in infiltrating T-cells, altogether promoting immune evasion (29). Notably, CREBBP mutations affecting the HAT domain confer a more severe functional phenotype (34). Functional genetic screens uncovered a synthetic lethal relationship between CREBBP and EP300, with CREBBP-mutant cancer cell lines showing a dependency on EP300 for their growth and survival by downregulating MYC expression (35,36).
The SET-domain histone methyltransferase, EZH2, is the enzymatic component of the polycomb repressor complex 2 (PRC2) and silences gene transcription by trimethylating histone 3 lysine 27 (H3K27me3). EZH2 mutations occur in approximately 20–25% of FL patients, with the majority of mutations clustered at 3 hotspot amino acid residues, Y641, A682 and A692 in the SET domain (37,38). The gain-of-function EZH2 mutations are heterozygous and promote an increase in H3K27 trimethylation. EZH2 appears essential for normal GC formation with mutant EZH2 regulating the GC phenotype-by suppressing cell cycle checkpoint genes such as CDKN1A and transcription factors that prevent GC exit such as IRF4 (39). Interestingly, recent studies have found EZH2 mutations also alter the tumour microenvironment (TME) by reducing tumour dependence on T-follicular helper (TFH) cells whilst shifting to follicular dendritic cell (FDC) interactions. By remodelling interactions with the microenvironment, B-cells no longer need to compete for TFH-dependent stimulation (which normally limits B-cell proliferation), enabling the large numbers of malignant-cells to persist in the GC (40). In DLBCL, MHC-I and MHC-II negative lymphomas are also strongly enriched for EZH2 mutations (41), supporting the additional role of EZH2 aberrations in immune evasion mechanisms.
In addition to mutations in histone-modifying genes, genes encoding components of chromatin remodelling complexes are also a feature of FL. Isoforms of the linker histones (HIST1H1B-E; also, H1B-E) are mutated in over 30% of FL tumours (42-44). These are predominantly heterozygous missense mutations clustered within the highly conserved globular domain, with the majority affecting the H1C and H1E isoforms. In normal cellular processes, these linker histones facilitate the folding of higher-order chromatin structures and regulate access of histone-modifying enzymes and chromatin remodelling complexes to their target genes (45). Aberrant H1C and H1E genes contribute to epigenetic reprogramming and gene silencing by impairing chromatin compaction and the 3D genome organisation thereby establishing H1 genes as tumour suppressors (42).
Most studies indicate that the mutations in epigenetic regulators, particularly CREBBP and KMT2D have high variant allelic fractions (VAFs) implying these mutations represent clonal genetic events. More remarkable is the co-existence of multiple ‘epimutations’ within a single FL tumour, at least 50% of cases harbour both KMT2D and CREBBP mutations, highlighting the importance of the convergence on H3K4 and H3K27, along the histone tail. The implications of this epigenetic intra-tumour heterogeneity and its phenotypic consequences have yet to be fully elucidated, although the nature of the histone marks suggests that the overall transcriptional effect is tipped preferentially towards a repressive gene expression state.
Mutations in immune modulators
Outside of epigenetic dysregulation, immune modulation is a mechanism frequently employed by tumours to evade the host’s natural immune responses. TNFRSF14 (also known as HVEM) is a bidirectional signalling molecule that interacts with its ligand, B and T lymphocyte attenuator (BTLA), to modulate T-cell activation. Mutations in TNFRSF14 occur in approximately 40% FLs, with the majority leading to loss of HVEM expression through gene deletions or truncations (27). TNFRSF14 aberrations disrupt the binding to its signalling partner, BTLA, and contributes to the generation of a tumour-supportive TME by increasing cytokines that promote TFH infiltration and activation of the tumour stroma (46-48). TNFRSF14 alterations have been linked with clinical outcome and increased GvHD risk following allogeneic stem cell transplant, although the data is somewhat conflicting and requires validation in larger series (27,47).
CTSS (Cathepsin S) encodes for a cysteine protease involved in MHC-II antigen presentation by antigen presenting cells (APCs) and malignant B-cells, regulating proteolytic cleavage of antigenic peptides and CD74. Mutations in CTSS occur in around 6% FLs, mainly by Y132D mutation resulting in CTSS overactivation, whilst CTSS overexpression is found in approximately 13% FLs (49,50). Enhanced CTSS activation in lymphomas increases antigen specific CD4+ T-cell activation and infiltration to garner tumour support and promote immune evasion through the exclusion of cytotoxic CD8+ T-cells (49,50). CTSS mutations and overexpression, which are mutually exclusive, also seem favourable when in patients treated with immunotherapies (50). Interestingly, CTSS Y132D mutations are mutually exclusive with TNFRSF14 and RRAGC (see below) mutations which alter CD4+ T-cell interactions (49).
Aberrations in mTORC1 signalling
Mutations converging on components of the amino-acid sensing arm of the mTORC1 signalling pathway have recently been reported (29,51,52). Recurrent mutations in the gene RRAGC, that encodes a Ras-related GTP-binding protein, Rag C, are enriched in FL, occurring in approximately 10–15% of patients, rarely present in other B-cell lymphomas. In vitro, RRAGC mutants can constitutively activate mTORC1 signalling even in amino acid deprived conditions suggesting that the mutant tumours have the capability of bypassing the normal metabolic checkpoint (51). RRAGC-mutant mice also have decreased tumour dependence on the microenvironment (53). Notably, RRAGC mutations frequently co-occur with mutations in ATP6V1B2 and ATP6AP1, subunits of the vacuolar ATPase proton pump, v-ATPase, a multimeric complex also needed for mTORC1 signalling (51). Another observation is the mutual exclusivity of RRAGC mutations with deletions in the gene Sestrin1 that encodes an upstream negative regulator of mTORC1. In aggregate, there are multiple genetic mechanisms that converge on mTORC1 signalling promoting aberrant metabolic reprogramming (54), emphasising its significance in FL pathogenesis.
Mutations in other signalling pathway components
Components of both the NF-κB and JAK-STAT pathway are subject to recurrent mutations in FL (43). Constitutive activation of the anti-apoptotic NF-κB signalling pathway, caused by mutations affecting positive and negative regulators, is an established feature of a number of B-cell lymphomas, particularly activated B-cell DLBCL (ABC-DLBCL). CARD11 encodes a key scaffolding protein in the CBM (CARD11-BCL10-MALT1) signalosome complex, which promotes NF-κB activation upon antigen receptor ligation in B-cells and is mutated in just over 10% of cases. TNFAIP3, encoding the enzyme A20 that acts as a negative regulator of canonical NF-κB signalling, is mutated at a similar frequency. CARD11 mutations occur within the coiled-coil domain and are activating aberrations (55), whilst the majority of the TNFAIP3 mutations are inactivating (56), usually combined with deletions of the second allele. Mutations in other components of the BCR- NF-κB signalling pathways including CD79A, CD79B and MYD88 are infrequently mutated in FL (52) compared to ABC-DLBCL or the newly recognised MCD/C5 DLBCL molecular subtypes (57,58).
In addition, mutations in the JAK-STAT signalling pathway, that is perhaps more synonymous with other lymphoma subtypes like Hodgkin lymphoma and primary mediastinal B-cell lymphoma are also recurrent in FL, leading to constitutive activation of the pathway and promoting B-cell survival (59). Activating mutations in STAT6 and inactivating mutations in SOCS1, a negative regulator of JAK-STAT signalling each occur in approximately 10% of FL cases. The activating STAT6 mutations induces a number of STAT6 target genes with the effects more pronounced in the presence of IL-4, a cytokine that independently drives STAT6 responsive genes, suggesting that both the mutations and the cytokine-driven signals from the microenvironment could activate the IL4-JAK-STAT axis supporting proliferation and survival of tumour cells (59).
Alterations affecting genes involved in proliferation and cell cycle regulation
Tumour cells co-opt oncogenes and circumvent tumour suppressors in order to proliferate uncontrollably and self-autonomously. In FL, several genes involved in the regulatory mechanisms of these processes are altered to give the tumour population a proliferative and survival advantage and are especially enriched at relapse and/or transformation (22,51,60-62). The proto-oncogene, MYC, can be mutated, amplified, or translocated in FL. Mutations and deletions of the genomic loci encompassing TP53, a tumour suppressor gene, occur in about 10–15% of FL, typically associated with adverse outcome. As alluded to earlier, certain cell cycle components and their regulators are recurrently perturbed by CNAs. The genomic region 12q13–15 is subject to frequent copy number gains and unsurprisingly this region encompasses several cell cycle regulatory components including MDM2, a ubiquitin-protein ligase that degrades TP53 and cyclin-dependent kinases (CDKs), CDK2 and CDK4, essential for G1/S transition of the cell cycle. CDK activity is negatively regulated by the inhibitor CDKN2A/p16, preventing their interaction with cyclin D and subsequent phosphorylation of the retinoblastoma (RB) proteins. The gene locus of CDKN2A/p16 has recurrent heterozygous and homozygous deletions, suggesting the removal of its inhibitory effect allows the potentially uncontrolled phosphorylation of RB proteins leading to the release of E2F transcription factors and continuous cell cycle progression. Cyclin D3 (CCND3), which encodes a binding partner of the CDKs, is mutated in about 5–10% of FL cases, co-occurring with CDK4 amplifications. Collectively, these alterations mostly occur in a mutually exclusive manner (63) and the perturbation of this set of genes converge on both the p53 and RB axis, decreasing their activity, deregulating the cell cycle and ultimately promoting tumour proliferation and survival.
In summary, there is a much clearer picture of the genetic, especially the mutational, landscape of FL and how these gene mutations deregulate specific biological pathways relevant for its pathogenesis. Whilst we have a better idea as to the degree of FL interpatient genetic heterogeneity, this has primarily been derived from, at times, small single centre studies on patient samples from a range of clinical phenotypes, as well as different modalities in genetic profiling from targeted gene panel to whole genome sequencing. From these studies, there are early insights into patterns of genetic co-dependencies and mutual exclusivity, although we are unaware if there are distinct genetic subtypes, as has been identified for example in DLBCL (57,58,64) which might explain the different FL clinical phenotypes. To address this, there is a need for much larger scale, statistically powered studies (65), to not only understand the contemporary pre-treatment genetic profiles but also allow us to capture underlying genetic subtypes and define how this may in turn influence disease evolution and clinical outcome.
Intra-tumour heterogeneity—temporal and spatial
There is increased recognition that tumours within individual patients consist of multiple genetically distinct subclones and that this intra-patient or intra-tumour heterogeneity can indeed act as the substrate for (sub)clonal evolution, treatment resistance and disease progression (66). The extent of this heterogeneity has really come into prominence by genetic profiling of tumours both temporally (where tumours at different clinical time points from an individual are examined, for example diagnostic versus relapse) and also spatially (tumours from different sites of disease at approximately the same time point).
In FL, temporal genetic analysis of sequential tumour samples (at diagnosis, relapse and/or transformed disease) have identified the patterns of clonal evolution (29,43,60-62). These studies demonstrated that the predominant pattern of clonal evolution was for relapsed or tFL tumours to arise via branched divergent clonal evolution. Here, the key observation also was that by using the genetic data to reconstruct the clonal phylogenies, sequential tumours appear to arise and diverge from a presumed ancestral population referred to as the common progenitor cell (CPC) that is shared across all the tumours from the same individual. The persistence of this ancestral CPC population through the patient’s clinical journey implies that this population is difficult to eradicate completely, especially with patients with recurrent disease (Figure 2). Further supporting evidence of the existence of a long-lived ancestral CPC population comes from two distinct cases of donor-derived FL (67,68). Here, both the donor and recipient develop FL many years after an allogeneic stem cell transplant with the tumours shown to be clonally related with identical t(14;18) translocations and other genetic events implying that an ancestral CPC must have been seeded at the time of the transplant and can exist for several years prior to overt FL development. Precise analyses of the genetic composition of temporally profiled tumours have brought into focus the early, initiating genetic events that must reside within this CPC population versus progression- or transformation-specific genetic aberrations. Mutations in the epigenetic regulators (CREBBP and KMT2D) together with the t(14;18) translocations represent initiating genetic events that drive lymphomagenesis, that must occur within the ancestral population and are predominantly conserved during the progression of the disease. Notably, some patients exhibited different mutations in CREBBP and KMT2D between FL and tFL, signifying convergent evolution and the importance of these mutations in lymphomagenesis (43,60).
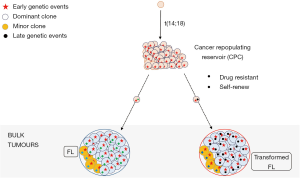
There is also an emphasis to understand the latter genetic events that contribute to progression and transformation. Early progression of FL after conventional chemotherapy and transformation to DLBCL (tFL) are both associated with inferior outcomes and represent one of the major causes of patient deaths from their lymphoma (2,3,69). Early progressed FLs appear to arise from an expansion of pre-existing subclones already present at diagnosis, indicating these subclones were to a degree resistant to initial therapy (61). Mutations in a number of genes including TP53, SOCS1, B2M and MYD88 were enriched in the diagnostic tumours of early progressed patients. Unsurprisingly, the genetic drivers of transformation are broad and heterogeneous with overlaps in gene mutations identified in both diagnostic and early progressed tumours, albeit seen at lower frequencies in pre-transformed biopsies. In addition to mutations in epigenetic regulators and the t(14;18) translocation that serve as early drivers in putative CPC populations, there are increased aberrant somatic hypermutation and aberrations in genes involved with cell cycle progression, proliferation and DNA damage response such as TP53, MYC, CDKN2A and REL in transformed samples (29,43,60,61). Increased immune escape mechanisms are also a feature of transformed disease with more frequent B2M mutations and deletions in parallel occurring with a reduced CD8+ T-cell infiltrate (61). The majority of tFLs retain a GCB-cell-of-origin gene expression signature however approximately 16% are classified as ABC-like (70). Interestingly, ABC-classified tFLs commonly evolve from t(14;18)-negative FL tumours and are associated with acquisition of NF-κβ mutations that are normally over-represented in ABC-DLBCLs (70). This coincides with a study demonstrating that an NF-κβ gene expression signature was associated with increased risk of transformation (71). This perhaps signals that NF-κβ biology may cultivate a more aggressive, fitter, subclonal phenotype in FL, as in DLBCL. The difference in GCB-like versus ABC-like tFLs did not appear to impact overall survival; although bearing in mind the caveats of the small heterogeneously-treated cohorts (70). Collectively, these longitudinal studies highlight the absence of a single (epi)genetic driver but instead, multifactorial genetic mechanisms enriched at transformation that contribute to the outgrowth and survival of more genomically complex subclones.
Spatial intra-tumour heterogeneity has been much less studied in FL but well reported in solid tumours (66). The majority of FL patients present with disease in multiple lymph nodes and other extra-nodal sites such as the bone marrow. By exome sequencing tumours from different sites of disease (spatially separated), there are expectedly varying degrees of spatial intra-tumour heterogeneity in FL (72). The mutations in CREBBP and KMT2D together with the t(14;18) translocation occur concordantly across spatially separated biopsies further emphasising their driver status in FL. However, the existence of inter-site heterogeneity is reminiscent of findings in other cancers and has important clinical implications for future biomarker-led therapeutic strategies. This is exemplified in a case that harbours an EZH2 mutation in a larger proportion of the tumour population in the lymph node compared to a small subclonal fraction in the bone marrow. Hypothetically, if this patient were treated with an EZH2 inhibitor such as tazemetostat, one might predict differential clearance of the tumour population at the two different sites of disease. There is a suggestion that this spatial heterogeneity increases at transformation (72). Recently, transcriptional heterogeneity across spatially-separated lymph nodes at the single cell level in FL patients has been demonstrated (73), suggesting that genetic diversity is just one of a multitude of layers that contribute to intra-tumour heterogeneity and that a single biopsy oversimplifies the molecular complexities of patients’ tumours.
Analysis of cell-free tumour DNA (ctDNA), fragments released into the blood, a means of liquid biopsy, may capture and provide a better representation of the patient’s intra-tumoral heterogeneity. Liquid biopsy analyses has been best studied in DLBCL with ctDNA able to capture the mutational landscape and clonal evolution as well as demonstrating prognostic relevance, with pre-treatment and interim ctDNA levels associated with outcome (74,75). Studies in FL are emerging with Delfau-Lareu and colleagues showing that ctDNA levels correlated with tumour burden and prognosis (76). Of even greater interest is if ctDNA has the utility to predict FL transformation. Scherer and colleagues showed, using CAPP-seq, that genetic events associated with transformation could be detected in the diagnostic ctDNA time point that was several months earlier (77). This minimally-invasive modality might offer the opportunity to capture heterogeneity whilst dynamically monitoring disease response to treatment and the ability to forecast progression. Further evaluations in this area are eagerly awaited.
Genetic heterogeneity between other FL-related entities
While it is clear that FL, tFL and DLBCL genetically overlap, an understanding of the trajectory from a normal B-cell to overt malignancy has been further improved by insights from a number of closely related entities, some recently described in the WHO classification 2016 update, with distinct clinico-pathological patterns compared to classical FL (78). This histological spectrum ranges from putative pre-malignant lesions like in situ follicular neoplasia (ISFN) to established malignancies such as t(14;18)-negative FL, paediatric-type FL (PTFL) and duodenal FL (DFL) (Table 1).

Full table
ISFN and partial involvement by FL (PFL)
ISFN, first identified in 2002, represents hyperplastic GCs colonised by CD10+ BCL2+ non-neoplastic B-cells (78-80). The true incidence of ISFN is unclear as they are typically detected inadvertently following biopsies for suspicious lymphadenopathy. It is reported to occur in approximately 2% of all individuals in which lymph nodes are removed for reasons other than lymphoma diagnostics (81). The risk of progression to overt classical FL is low (<5%) suggesting these presumed precursor lesions are yet to have acquired the full complement of genetic changes for lymphomagenesis. Unlike ISFN, PFL more closely resembles overt FL with altered lymph node architecture and higher rates of progression, possibly representing a more advanced pre-malignant stage (80,82). However half of patients with PFL still do not undergo malignant transformation (80,82,83). Notably, common genetic alterations seen in overt FL including mutations in EZH2, KMT2D and TNFRSF14, were identified in both ISFN and PFL, further supporting evidence for their early driver status (84). The degree of genomic complexity (CNAs and gene mutations) increased comparatively from ISFN to PFL to overt FL, suggesting a potential evolutionary hierarchy within these entities toward FL development (84).
T(14;18)-negative FL
T(14;18)-negative FL represents 10–15% of FLs and is more prevalent in early stage FL, which has a superior prognosis (85). Although lacking the BCL2 translocation, t(14;18)-negative FL patients exhibit similar clinical features to their t(14;18)-positive counterparts and the majority still express BCL2 protein (86,87). Whilst t(14;18)-negative FLs share much of the CNA profile of conventional FL, these occur at lower frequencies (85). Interestingly, the gene expression profile in t(14;18)-negative FLs are more reminiscent of ABC-like B-cell tumours with a particular enrichment for NF-κB signatures (87) and perhaps explains our earlier description that ABC-like tFLs more often evolve from a preceding t(14;18)-negative FL. STAT6 mutations are also more prevalent compared to conventional FL (57% vs. 12%) (85) and frequently co-occur with CREBBP or TNFRSF14 mutations, emphasising the alternative oncogenic potential even in the absence of the BCL2 translocation. There is no suggestion that t(14;18)-negative FLs need to be managed any differently to conventional FL.
DFL
DFL is a recognised variant anatomically restricted to the duodenum that follows a relatively benign clinical course and is morphologically similar to FL. Genetically, DFLs harbour mutations in genes seen in classical FL including TNFRSF14, EZH2 and CREBBP, however have a significantly lower frequency of KMT2D mutations and extremely rare progression to overt FL (84,88-91). Unlike overt FL where activation-induced cytidine deaminase (AID) is highly expressed, consistent with its GC origin, DFLs rarely express AID—hinting DFLs originate from a non-GC B-cell that migrated to the duodenum (89). A particularly differentiating feature of DFL is their distinct immune microenvironment compared to classical FL, with a gene expression signature of chronic inflammation that may also contribute to the clinical and anatomical differences between the other FL entities (91).
PTFL
PTFL is a variant of FL presenting as localized lymphadenopathy in the head and neck with a male preponderance that occurs mostly in children but also in adults. PTFLs lack the t(14;18) signature lesion seen in classical FL but have a surprisingly high proliferation index (>30%) for a disease with a typically excellent prognosis and low rates of recurrence (92). The genomic complexity of PTFLs is low with few CNA, an enrichment in mutations in TNFRSF14 and genes involved in the MAPK pathway (such as MAP2K1, as high as 40%) but a paucity of mutations in epigenetic regulators (93,94), altogether supporting different routes of disease initiation compared to both classical and t(14;18)-negative FL.
Translational relevance of FL genetics
We now stand at a crossroads with the deluge of genomic information in FL and how best to prioritise and meaningfully translate this knowledge into patient benefit. Given the areas of unmet need in FL, potential avenues include development of targeted therapies directed to aberrant genomic profiles and improved biomarkers for risk stratification, disease monitoring and therapeutic response.
New therapeutic targets
As mutations in the epigenetic machinery are frequent and early driver events, there is much focus in developing therapeutic opportunities to reverse the impact of these mutations. The gain-of-function nature of EZH2 mutations have made them particularly attractive targets with several direct small-molecule EZH2 inhibitors already developed and being evaluated in clinical trials, including tazmetostat (95). In a phase II study of relapsed/refractory FL treated with tazmetostat, patients with EZH2-mutant lymphomas had superior overall response (96) compared to EZH2 wild-type. Interestingly, tazmetostat also rescues MHC expression and restores T-cell infiltration in EZH2-mutant-cell lines and mouse models (41), abrogating the immune evasion effects of the mutation. This could open the door to evaluating combinations of EZH2 inhibition with immunotherapies to enhance immune recognition and synergistically potentiate the efficacy of these therapies.
Potential therapeutics specifically targeting CREBBP mutant lymphomas are gathering momentum. CREBBP loss-of-function in FL leads to HDAC3-mediated suppression of gene enhancers. Although, pan-HDAC inhibitors have shown limited activity in B-cell lymphomas in early-phase studies (97,98), selective HDAC3-inhibitors may offer a more direct approach to counteracting CREBBP mutations (32,34,99). The synthetic lethality between CREBBP and EP300 may also be exploited as CREBBP-mutant cell lines showed more susceptibility to deletion of EP300 or pharmacological inhibition with HAT or bromodomain inhibitors (35,36). CCS1477, a first-in-class small molecule inhibitor of the p300/CBP conserved bromodomain (100) is currently being evaluated in early phase clinical trials in haematological malignancies.
Genomics informing predictive and prognostic markers
Identifying patients with high-risk FL is an area of unmet clinical need. Historically clinical information has been the bedrock for prognostic tools, although did not influence treatment decisions (4,5). Genomic information is now being integrated into prognostic tools such as the m7-FLIPI index, which assesses the mutation status of seven genes (EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, CARD11) together with clinical characteristics to risk stratify patients (101). Whilst the prognostic accuracy of the m7-FLIPI model may be treatment-dependent and not necessarily capture all high-risk patients, suggesting that different mutations may be implicated in response and resistance to different therapies. Nonetheless, it is an important stepping-stone to incorporating molecular parameters into risk stratification tools (8,102). A more recent prognostic iteration developed by Huet and colleagues (7) uses the expression of 23 genes, encompassing B-cell biology and the TME, to identify patients at increased risk of progression.
With increasing therapeutic options in FL in the first line and relapsed settings, defining biological predictors of both therapeutic response and resistance will be invaluable to select therapy. EZH2 mutations already serve as a good predictive biomarker of response to the EZH2 inhibitor, tazemetostat. A recent, retrospective analysis of the phase III GALLIUM trial (96) identified a predictive link between EZH2 mutations, and the chemotherapy backbone used to treat FL patients in the first line setting. EZH2-mutated FL patients had better clinical outcomes with a CHOP/CVP backbone compared to EZH2 wild-type patients, irrespective of anti-CD20 therapy, suggesting that genetic mutations could also influence how we use conventional therapies, although this needs further evaluation (102). One anticipates that we will start to see many such studies to identify appropriate and accurate predictive biomarkers of response to conventional and novel therapies including immunotherapies such as CAR-T.
Future perspective and conclusions
The last decade has illuminated the breadth of genomic complexity and an appreciation of the heterogeneity in FL that a single biopsy, whilst informative, inadequately captures. The spatial and temporal heterogeneity may undermine accurate prognostication and impact on mutation-specific treatments. The residual CPC population potentiates the recurrent relapse-remitting nature of FL and could represent the Achilles’ heel of these tumours. It is unclear if such CPC cells reside within the minimal residual disease (MRD) population that persist after treatment and the next focus is to understand the nature of the FL CPC: what are its characteristics, is there heterogeneity within this CPC and does this relate to the clinical phenotype and evolution of the disease.
Each FL tumour is a compendium of several genetically distinct subclones dependent on different epigenetic and signalling pathways, therefore therapeutically targeting a single genetic aberration is unlikely to be a successful long-term strategy in every patient as it would enhance subclonal competition and promote the outgrowth of resistant clones. Although the genetic and epigenetic signatures and heterogeneity are key drivers of this disease, other components such as the TME also play a supporting role. Combinatorial therapies targeting multiple tumour vulnerabilities coupled with means of measuring the response and clonal dynamics, for example with ctDNA assays, may prove to be the most effective strategy.
Acknowledgments
Funding: JO is funded by Cancer Research UK (C57432/A22742) and MP is supported by a Cancer Research UK Accelerator Award Studentship (C355/A28222).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Mark Roschewski, Carla Casulo) for the series “Follicular Lymphoma” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-21-5). The series “Follicular Lymphoma” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jurinovic V, Kridel R, Staiger AM, et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood 2016;128:1112-20. [Crossref] [PubMed]
- Casulo C, Byrtek M, Dawson KL, et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the National LymphoCare Study. J Clin Oncol 2015;33:2516-22. [Crossref] [PubMed]
- Wagner-Johnston ND, Link BK, Byrtek M, et al. Outcomes of transformed follicular lymphoma in the modern era: a report from the National LymphoCare Study (NLCS). Blood 2015;126:851-7. [Crossref] [PubMed]
- Solal-Céligny P, Roy P, Colombat P, et al. Follicular lymphoma international prognostic index. Blood 2004;104:1258-65. [Crossref] [PubMed]
- Federico M, Bellei M, Marcheselli L, et al. Follicular lymphoma international prognostic index 2: a new prognostic index for follicular lymphoma developed by the international follicular lymphoma prognostic factor project. J Clin Oncol 2009;27:4555-62. [Crossref] [PubMed]
- Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015;16:1111-22. [Crossref] [PubMed]
- Huet S, Tesson B, Jais JP, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol 2018;19:549-61. [Crossref] [PubMed]
- Lockmer S, Ren W, Brodtkorb M, et al. M7-FLIPI is not prognostic in follicular lymphoma patients with first-line rituximab chemo-free therapy. Br J Haematol 2020;188:259-67. [Crossref] [PubMed]
- Araf S, Okosun J, Fitzgibbon J. Predicting early relapse in follicular lymphoma: have we turned a corner? Lancet Oncol 2018;19:441-2. [Crossref] [PubMed]
- Cleary ML, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci U S A 1985;82:7439-43. [Crossref] [PubMed]
- Yunis JJ, Frizzera G, Oken MM, et al. Multiple recurrent genomic defects in follicular lymphoma. N Engl J Med 1987;316:79-84. [Crossref] [PubMed]
- Cleary ML, Galili N, Sklar J. Detection of a second t(14;18) breakpoint cluster region in human follicular lymphomas. J Exp Med 1986;164:315-20. [Crossref] [PubMed]
- Ngan BY, Nourse J, Cleary ML. Detection of chromosomal translocation t(14;18) within the minor cluster region of bcl-2 by polymerase chain reaction and direct genomic sequencing of the enzymatically amplified DNA in follicular lymphomas. Blood 1989;73:1759-62. [Crossref] [PubMed]
- Akasaka T, Akasaka H, Yonetani N, et al. Refinement of the BCL2/immunoglobulin heavy chain fusion gene in t(14;18)(q32;q21) by polymerase chain reaction amplification for long targets. Genes Chromosom Cancer 1998;21:17-29. [Crossref] [PubMed]
- Willis TG, Jadayel DM, Coignet LJ, et al. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood 1997;90:2456-64. [Crossref] [PubMed]
- Buchonnet G, Lenain P, Ruminy P, et al. Characterisation of BCL2-JH rearrangements in follicular lymphoma: PCR detection of 3’ BCL2 breakpoints and evidence of a new cluster. Leukemia 2000;14:1563-9. [Crossref] [PubMed]
- Albinger-Hegyi A, Hochreutener B, Abdou MT, et al. High frequency of t(14;18)-translocation breakpoints outside of major breakpoint and minor cluster regions in follicular lymphomas: improved polymerase chain reaction protocols for their detection. Am J Pathol 2002;160:823-32. [Crossref] [PubMed]
- Limpens J, Stad R, Vos C, et al. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood 1995;85:2528-36. [Crossref] [PubMed]
- Dölken G, Illerhaus G, Hirt C, et al. BCL-2/JH rearrangements in circulating b cells of healthy blood donors and patients with nonmalignant diseases. J Clin Oncol 1996;14:1333-44. [Crossref] [PubMed]
- Schüler F, Dölken L, Hirt C, et al. Prevalence and frequency of circulating (14;18)-MBE translocation carrying cells in healthy individuals. Int J Cancer 2009;124:958-63. [Crossref] [PubMed]
- Roulland S, Kelly RS, Morgado E, et al. t(14;18) translocation: a predictive blood biomarker for follicular lymphoma. J Clin Oncol 2014;32:1347-55. [Crossref] [PubMed]
- Bouska A, McKeithan TW, Deffenbacher KE, et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014;123:1681-90. [Crossref] [PubMed]
- Cheung KJJ, Shah SP, Steidl C, et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood 2009;113:137-48. [Crossref] [PubMed]
- Viardot Aa, Barth TF, Möller P, et al. Cytogenetic evolution of follicular lymphoma. Semin Cancer Biol 2003;13:183-90. [Crossref] [PubMed]
- Viardot A, Möller P, Högel J, et al. Clinicopathologic correlations of genomic gains and losses in follicular lymphoma. J Clin Oncol 2002;20:4523-30. [Crossref] [PubMed]
- Johnson NA, Al-Tourah A, Brown CJ, et al. Prognostic significance of secondary cytogenetic alterations in follicular lymphomas. Genes Chromosomes Cancer 2008;47:1038-48. [Crossref] [PubMed]
- Cheung KJJ, Johnson NA, Affleck JG, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res 2010;70:9166-74. [Crossref] [PubMed]
- Oricchio E, Nanjangud G, Wolfe AL, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell 2011;147:554-64. [Crossref] [PubMed]
- Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci 2015;112:E1116-25. [Crossref] [PubMed]
- Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21:1190-8. [Crossref] [PubMed]
- Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med 2015;21:1199-208. [Crossref] [PubMed]
- Jiang Y, Ortega-Molina A, Geng H, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov 2017;7:38-53. [Crossref] [PubMed]
- Zhang J, Vlasevska S, Wells VA, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov 2017;7:322-37. [Crossref] [PubMed]
- Mondello P, Tadros S, Teater M, et al. Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov 2020;10:440-59. [Crossref] [PubMed]
- Meyer SN, Scuoppo C, Vlasevska S, et al. Unique and shared epigenetic programs of the CREBBP and EP300 acetyltransferases in germinal center B cells reveal targetable dependencies in lymphoma. Immunity 2019;51:535-47.e9. [Crossref] [PubMed]
- Ogiwara H, Sasaki M, Mitachi T, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov 2016;6:430-45. [Crossref] [PubMed]
- Bödör C, Grossmann V, Popov N, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 2013;122:3165-8. [Crossref] [PubMed]
- Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010;42:181-5. [Crossref] [PubMed]
- Béguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013;23:677-92. [Crossref] [PubMed]
- Béguelin W, Teater M, Meydan C, et al. Mutant EZH2 induces a pre-malignant lymphoma niche by reprogramming the immune response. Cancer Cell 2020;37:655-73.e11. [Crossref] [PubMed]
- Ennishi D, Takata K, Béguelin W, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov 2019;9:546-63. [Crossref] [PubMed]
- Yusufova N, Kloetgen A, Teater M, et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 2021;589:299-305. [Crossref] [PubMed]
- Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 2014;46:176-81. [Crossref] [PubMed]
- Li H, Kaminski MS, Li Y, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood 2014;123:1487-98. [Crossref] [PubMed]
- Fyodorov DV, Zhou BR, Skoultchi AI, et al. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 2018;19:192-206. [Crossref] [PubMed]
- Kotsiou E, Okosun J, Besley C, et al. TNFRSF14 aberrations in follicular lymphoma increase clinically significant allogeneic T-cell responses. Blood 2016;128:72-81. [Crossref] [PubMed]
- Launay E, Pangault C, Bertrand P, et al. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia 2012;26:559-62. [Crossref] [PubMed]
- Boice M, Salloum D, Mourcin F, et al. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell 2016;167:405-18.e13. [Crossref] [PubMed]
- Dheilly E, Battistello E, Katanayeva N, et al. Cathepsin S regulates antigen processing and T cell activity in non-Hodgkin lymphoma. Cancer Cell 2020;37:674-89.e12. [Crossref] [PubMed]
- Bararia D, Hildebrand JA, Stolz S, et al. Cathepsin S alterations induce a tumor-promoting immune microenvironment in follicular lymphoma. Cell Rep 2020;31:107522 [Crossref] [PubMed]
- Okosun J, Wolfson RL, Wang J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48:183-8. [Crossref] [PubMed]
- Krysiak K, Gomez F, White BS, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 2017;129:473-83. [Crossref] [PubMed]
- Ortega-Molina A, Deleyto-Seldas N, Carreras J, et al. Oncogenic Rag GTPase signalling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Nat Metab 2019;1:775-89. [Crossref] [PubMed]
- Oricchio E, Katanayeva N, Donaldson MC, et al. Genetic and epigenetic inactivation of SESTRIN1 controls mTORC1 and response to EZH2 inhibition in follicular lymphoma. Sci Transl Med 2017;9:eaak9969 [Crossref] [PubMed]
- Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008;319:1676-9. [Crossref] [PubMed]
- Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood 2009;114:2467-75. [Crossref] [PubMed]
- Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396-407. [Crossref] [PubMed]
- Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679-90. [Crossref] [PubMed]
- Yildiz M, Li H, Bernard D, et al. Lymphoid neoplasia: activating stat6 mutations in follicular lymphoma. Blood 2015;125:668-79. [Crossref] [PubMed]
- Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep 2014;6:130-40. [Crossref] [PubMed]
- Kridel R, Chan FC, Mottok A, et al. Histological transformation and progression in follicular lymphoma: a clonal evolution study. PLoS Med 2016;13:e1002197 [Crossref] [PubMed]
- Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood 2013;121:1604-11. [Crossref] [PubMed]
- Oricchio E, Ciriello G, Jiang M, et al. Frequent disruption of the RB pathway in indolent follicular lymphoma suggests a new combination therapy. J Exp Med 2014;211:1379-91. [Crossref] [PubMed]
- Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood 2020;135:1759-71. [Crossref] [PubMed]
- Li X, Kositsky R, Reddy A, et al. Whole exome and transcriptome sequencing in 1042 cases reveals distinct clinically relevant genetic subgroups of follicular lymphoma. Blood 2019;134:19. [Crossref]
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Carlotti E, Wrench D, Matthews J, et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma may occur by divergent evolution from a common progenitor cell or by direct evolution from the follicular lymphoma clone. Blood 2009;113:3553-7. [Crossref] [PubMed]
- Weigert O, Kopp N, Lane AA, et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov 2012;2:47-55. [Crossref] [PubMed]
- Sarkozy C, Maurer MJ, Link BK, et al. Cause of death in follicular lymphoma in the first decade of the rituximab era: a pooled analysis of French and US cohorts. J Clin Oncol 2019;37:144-52. [Crossref] [PubMed]
- Kridel R, Mottok A, Farinha P, et al. Cell of origin of transformed follicular lymphoma. Blood 2015;126:2118-27. [Crossref] [PubMed]
- Brodtkorb M, Lingjærde OC, Huse K, et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood 2014;123:1051-4. [Crossref] [PubMed]
- Araf S, Wang J, Korfi K, et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2018;32:1261-5. Erratum in: Leukemia 2019;33:1540. [Crossref] [PubMed]
- Haebe S, Shree T, Sathe A, et al. Site to site comparison of follicular lymphoma biopsies by single cell RNA sequencing. Blood 2019;134:297. [Crossref]
- Kurtz DM, Scherer F, Jin MC, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 2018;36:2845-53. [Crossref] [PubMed]
- Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 2015;16:541-9. [Crossref] [PubMed]
- Delfau-Larue MH, Van Der Gucht A, Dupuis J, et al. Total metabolic tumor volume, circulating tumor cells, cell-free DNA: distinct prognostic value in follicular lymphoma. Blood Adv 2018;2:807-16. [Crossref] [PubMed]
- Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016;8:364ra155 [Crossref] [PubMed]
- Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375-90. [Crossref] [PubMed]
- Cong P, Raffeld M, Teruya-Feldstein J, et al. In situ localization of follicular lymphoma: description and analysis by laser capture microdissection. Blood 2002;99:3376-82. [Crossref] [PubMed]
- Jegalian AG, Eberle FC, Pack SD, et al. Follicular lymphoma in situ: clinical implications and comparisons with partial involvement by follicular lymphoma. Blood 2011;118:2976-84. [Crossref] [PubMed]
- Henopp T, Quintanilla-Martínez L, Fend F, et al. Prevalence of follicular lymphoma in situ in consecutively analysed reactive lymph nodes. Histopathology 2011;59:139-42. [Crossref] [PubMed]
- Pillai RK, Surti U, Swerdlow SH. Follicular lymphoma-like B cells of uncertain significance (in situ follicular lymphoma) may infrequently progress, but precedes follicular lymphoma, is associated with other overt lymphomas and mimics follicular lymphoma in flow cytometric studies. Haematologica 2013;98:1571-80. [Crossref] [PubMed]
- Mamessier E, Broussais-Guillaumot F, Chetaille B, et al. Nature and importance of follicular lymphoma precursors. Haematologica 2014;99:802-10. [Crossref] [PubMed]
- Mamessier E, Song JY, Eberle FC, et al. Early lesions of follicular lymphoma: a genetic perspective. Haematologica 2014;99:481-8. [Crossref] [PubMed]
- Nann D, Ramis-Zaldivar JE, Müller I, et al. Follicular lymphoma t(14;18)-negative is genetically a heterogeneous disease. Blood Adv 2020;4:5652-65. [Crossref] [PubMed]
- Leich E, Hoster E, Wartenberg M, et al. Similar clinical features in follicular lymphomas with and without breaks in the BCL2 locus. Leukemia 2016;30:854-60. [Crossref] [PubMed]
- Leich E, Salaverria I, Bea S, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood 2009;114:826-34. [Crossref] [PubMed]
- Bende RJ, Smit LA, Bossenbroek JG, et al. Primary follicular lymphoma of the small intestine: α4β7 expression and immunoglobulin configuration suggest an origin from local antigen-experienced B cells. Am J Pathol 2003;162:105-13. [Crossref] [PubMed]
- Takata K, Sato Y, Nakamura N, et al. Duodenal follicular lymphoma lacks AID but expresses BACH2 and has memory B-cell characteristics. Mod Pathol 2013;26:22-31. Erratum in: Mod Pathol 2013;26:1152. [Crossref] [PubMed]
- Schmatz AI, Streubel B, Kretschmer-Chott E, et al. Primary follicular lymphoma of the duodenum is a distinct mucosal/submucosal variant of follicular lymphoma: A retrospective study of 63 cases. J Clin Oncol 2011;29:1445-51. [Crossref] [PubMed]
- Hellmuth JC, Louissaint A, Szczepanowski M, et al. Duodenal-type and nodal follicular lymphomas differ by their immune microenvironment rather than their mutation profiles. Blood 2018;132:1695-702. [Crossref] [PubMed]
- Louissaint A, Ackerman AM, Dias-Santagata D, et al. Pediatric-type nodal follicular lymphoma: an indolent clonal proliferation in children and adults with high proliferation index and no BCL2 rearrangement. Blood 2012;120:2395-404. [Crossref] [PubMed]
- Louissaint A, Schafernak KT, Geyer JT, et al. Pediatric-type nodal follicular lymphoma: a biologically distinct lymphoma with frequent MAPK pathway mutations. Blood 2016;128:1093-100. [Crossref] [PubMed]
- Schmidt J, Gong S, Marafioti T, et al. Genome-wide analysis of pediatric-type follicular lymphoma reveals low genetic complexity and recurrent alterations of TNFRSF14 gene. Blood 2016;128:1101-11. [Crossref] [PubMed]
- Knutson SK, Kawano S, Minoshima Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther 2014;13:842-54. [Crossref] [PubMed]
- Morschhauser F, Tilly H, Chaidos A, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol 2020;21:1433-42. [Crossref] [PubMed]
- Kirschbaum M, Frankel P, Popplewell L, et al. Phase II study of vorinostat for treatment of relapsed or refractory indolent non-hodgkin’s lymphoma and mantle cell lymphoma. J Clin Oncol 2011;29:1198-203. [Crossref] [PubMed]
- Ogura M, Ando K, Suzuki T, et al. A multicentre phase II study of vorinostat in patients with relapsed or refractory indolent B-cell non-Hodgkin lymphoma and mantle cell lymphoma. Br J Haematol 2014;165:768-76. [Crossref] [PubMed]
- Höpken UE. Targeting HDAC3 in CREBBP -mutant lymphomas counterstrikes unopposed enhancer deacetylation of B-cell signaling and immune response genes. Cancer Discov 2017;7:14-6. [Crossref] [PubMed]
- Pegg N, Brooks N, Worthington J, et al. Characterisation of CCS1477: a novel small molecule inhibitor of p300/CBP for the treatment of castration resistant prostate cancer. J Clin Oncol 2017;35:11590. [Crossref]
- Vindi J, Kridel R, Staiger AM, et al. A clinicogenetic risk model (m7-FLIPI) prospectively identifies one-half of patients with early disease progression of follicular lymphoma after first-line immunochemotherapy. Blood 2015;126:333. [Crossref]
- Jurinovic V, Passerini V, Oestergaard MZ, et al. Evaluation of the m7-FLIPI in patients with follicular lymphoma treated within the gallium trial: EZH2 mutation status may be a predictive marker for differential efficacy of chemotherapy. Blood 2019;134:122. [Crossref]
Cite this article as: Perrett M, Okosun J. Genetic heterogeneity in follicular lymphoma. Ann Lymphoma 2021;5:18.


