The contribution of ebv to the pathogenesis of classical hodgkin lymphoma
Introduction
The oncogenic Epstein-Barr virus (EBV) is present in a subset of cases of classical Hodgkin lymphoma (cHL). Despite growing evidence of its importance in the pathogenesis of the subset of virus-associated cHL, therapies that specifically target EBV are lacking. We believe it is now timely to revisit the contribution of EBV to the pathogenesis of cHL, particularly with respect to the role of the viral latent genes, some of which are now being explored as targets of new drug and immunotherapeutic approaches.
General features and pathogenesis of HL
Hodgkin lymphoma (HL) has an incidence of around 3/100,000 per year. Lymph nodes are the most often affected tissues, although extranodal disease does occur. Affected tissues show effacement of existing structures which are replaced by rare malignant cells, surrounded by a florid reactive infiltrate. Based on differences in histology and immune phenotype, HL can be classified into two major types; these are classical HL (cHL) and nodular lymphocyte predominant (NLP) HL (1). The malignant cells of cHL are known as Hodgkin/Reed-Sternberg (HRS) cells, and those of NLPHL as lymphocyte predominant (LP) cells. There are four major subtypes of cHL, known as mixed cellularity, nodular sclerosis, lymphocyte rich and lymphocyte depleted HL. The tumour cells of HL are derived from mature B lymphocytes (2), and display evidence of somatic hypermutation indicating that they develop from germinal centre, or more likely, post-germinal centre B cells (2-5). In approximately one-quarter of cases of cHL, the immunoglobulin genes have so called ‘crippling’ mutations which prevent proper expression of surface immunoglobulin (3). In fact it is now believed that the loss of B cell receptor (BCR) functions is probably directly involved in the pathogenesis of most, if not all, cases of cHL.
Most B cells require signalling through the BCR for their survival, yet HRS cells do not appear to need these survival signals. These observations suggest that a crucial event must be the acquisition of mechanisms that prevent the apoptosis that would be the normal fate of germinal centre B cells lacking a functional BCR. Multiple cell signalling pathways are aberrantly activated in HRS cells, many of which contribute to this anti-apoptotic phenotype. For example, HRS cells display constitutive activation of a family of transcription factors known as nuclear factor kappa B (NF-κB) (6). Inhibition of NF-κB signalling in HL cell lines increases their sensitivity to apoptosis and impairs tumourigenicity in severe combined immunodeficiency mice (7,8). HRS cells express multiple tumour necrosis factor receptors, including CD30, CD40, TACI, BCMA and RANK which can induce the activation of NF-κB signalling following their engagement with ligands expressed on immune and other cells of the tumour microenvironment (TME) (9,10). Constitutive NF-κB activation can also be caused by different genetic lesions in HRS cells, including amplification of the gene encoding the c-REL subunit of NF-κB, (11-13), mutations in genes encoding inhibitors, IκB alpha and IκB epsilon (14-18), and TNFAIP3/A20, a ubiquitin modifying enzyme that inhibits NF-κB signalling (19).
The JAK/STAT signalling pathway is also critically involved in cHL pathogenesis and may be activated via the cytokines which are produced either by HRS cells or by cells of the TME; this in turn elevates the levels of phosphorylated forms of STATs (e.g., STAT3, STAT5A and STAT6) (20-22). JAK/STAT signalling can also be aberrantly stimulated by genetic lesions such as the amplification of JAK2 or loss-of-function mutations in SOCS1 and PTPN1/PTPB1 which are negative regulators of these pathways (23-25).
Identification of EBV in cHL
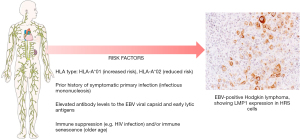
EBV was initially suggested to be involved in the pathogenesis of cHL after it was shown that patients had raised antibody levels to EBV antigens in their blood (26). Moreover, elevated levels were also shown to be present in patients before the onset of cHL (27).
Southern blotting for EBV DNA was first used to identify the presence of EBV DNA in the tissues of cHL patients (28). EBV was localized to HRS cells using the anti-complement immune fluorescence assay (29), by in situ hybridization (ISH) for EBV DNA (30,31), and by ISH for two RNA species known as Epstein-Barr virus encoded RNAs (EBER1 and EBER2) (32). An aetiological role for EBV in cHL was supported by the detection of viral genomes bearing identical fusion sequences in biopsies. These sequences are created when the circular genomes are formed from linear virus DNA and are unique to a single infection event. Thus, monoclonal viral genomes present in cHL indicate that infection is an early event (30). EBV is consistently retained during disease progression, suggesting it is required for maintenance of the tumour phenotype (33).
Epidemiology of EBV-associated cHL
EBV is associated with cHL, but not with NLPHL. However, not all cases of cHL are EBV-positive. Moreover, the proportion of positive cases varies within different populations (34,35). Thus, in resource-rich nations, EBV-positive rates range between 20% to 50%; here more EBV-positive cases are seen in older people and in children, and fewer in young adults (36,37). In contrast, EBV positive rates are often substantially higher in resource-poor countries (38,39). EBV-positive disease is more common in males, in patients with mixed cellularity disease, and in some ethnic groups, even when taking into account potential confounding factors, such as socioeconomic status (40).
The incidence of EBV-positive cHL is also more common in HIV-positive individuals especially when the levels of immune impairment are at intermediate levels (41,42). Thus, declining levels of EBV-specific immunity in the early stages of HIV infection contribute to an increased risk of EBV-positive HL. However, this risk of developing cHL declines as CD4+ T cell numbers fall further, emphasising the importance of CD4+ T cells in the pathogenesis of cHL.
First degree relatives of patients with cHL patients have between a 3-fold and 9-fold increased risk of developing the disease (43,44), this rises to 100-fold for monozygotic twins compared with dizygotic twins (45). Susceptibility loci exist within the human leukocyte antigen (HLA) region (46,47); HLA-A*01 and HLA-A*02 alleles confer an increased and decreased risk of EBV-positive cHL, respectively (47-49) (Figure 1). These findings illustrate the potential importance of immune control of the virus in the pathogenesis of the EBV-positive form of cHL (50). A prior history of infectious mononucleosis (IM) is associated with an increased risk of developing EBV-positive, but not EBV-negative, cHL (51-53) and some of the same HLA associations are also observed for IM (Figure 1).
EBV potently transforms B lymphocytes in vitro
Before considering how EBV might contribute to the development of cHL, we briefly outline what is known about the transforming potential of EBV and how the virus might interact with B cells in the normal asymptomatic host.
Infection of resting B lymphocytes by EBV in vitro can lead to their transformation, giving rise to continuously growing cell lines, referred to as lymphoblastoid cell lines (LCL). If lymphocytes from the blood are used, the T lymphocytes present must be depleted or suppressed by drugs such as cyclosporin A (54), demonstrating the importance of the T cell response in controlling EBV infection in vivo.
EBV encodes a small subset of its genes in the latency phase of the virus life cycle which is characterized by the absence of virus replication. These ‘latent’ genes include nuclear antigens, known as EBNAs (EBNAs 1, 2, 3A, 3B, 3C, EBNA-LP), latent membrane proteins (LMP1, LMP2), as well as two non-coding Epstein-Barr-encoded RNA (EBER1, EBER2), and viral miRNA (55,56). Some latent genes, for example, EBNA2 and LMP1 are necessary for the transformation of B cells, at least in the laboratory (57). Latency III is the term used to describe the type of latency displayed by LCL; here all known latent genes are present. Restricted forms of latency are observed during the ‘normal’ differentiation of EBV-infected B cells in vivo, as well as in EBV-associated malignancies, including cHL. EBV can also express ‘lytic’ genes when the virus shifts to its replicative cycle ending in the release of infectious virus particles.
EBV infection of B cells is characteristic of the asymptomatic carrier state
EBV has evolved to survive for the life-time of the asymptomatic host, and does so using a highly evolved mechanism that relies on the colonisation of memory B cells (58). How EBV eventually gets into memory B cells has been difficult to fully define, mainly because primary infection itself largely goes unnoticed and when IM does occur after primary infection, by the time it becomes symptomatic, the early events in virus infection have already taken place. To further exacerbate an already difficult problem, the virus colonises only a minute fraction of B cells in asymptomatic carriers (perhaps as low as 1 in 106 B cells). Notwithstanding these difficulties, the favoured model of EBV persistence suggests that initial infection of B cells by EBV causes them to enter the cell cycle, and to express the latency III pattern of viral gene expression; in this respect the initially infected B cells are probably similar to LCL. These EBV-infected B cells may then adopt a phenotype more closely resembling a germinal centre (GC) B cell, this time they adopt a latency II programme in which EBNA1, but not the other EBNAs, and the latent membrane proteins are present (59). This pattern of virus expression mirrors that seen in cHL. At this stage, LMP1 and LMP2 provide the EBV-infected B cells with surrogate CD40 and B cell receptor (BCR) signals, respectively, and allow the EBV-infected GC B cells to survive and subsequently to differentiate into memory B cells (60,61). The EBV-infected memory B cells no longer express virus genes (a state known as latency 0), but can if necessary switch on EBNA1 expression to allow them to proliferate (this stage is known as latency I) (59). EBV-infected B cells can become plasma cells; in this case the lytic cycle is induced, eventually leading to virion release in oropharyngeal secretions. This is probably the major way in which the virus to transmitted from host to host (62).
It is important to also mention here that there is an alternative view of the establishment of the carrier state which is based on the observation that EBV-infected cells in GCs of patients with IM show evidence of somatic hypermutation, but without evidence of intra-clonal diversity; this means that these cells are no longer undergoing the hypermutation process (63,64). Moreover, LMP1 has been shown to be present in EBV-infected cells present outside the GC, potentially counteracting the argument that LMP1 is driving a GC reaction (65). Thus, this alternative model proposes that EBV directly infects memory B cells. Moreover, ‘non-switched’ memory B cells, which do not require GC activity can also harbour EBV (66-68). EBV infection can induce expression of activation-induced cytosine deaminase (AID) to stimulate somatic hypermutation (69).
EBV latent genes are important contributors to the pathogenesis of cHL
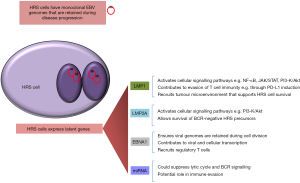
EBV-infected HRS cells express a restricted pattern of virus latency characterised by the presence of EBV’s maintenance protein, EBNA1, as well as both latent membrane proteins. A subset of viral miRNA are also expressed. While the contribution of the EBV latent proteins to the pathogenesis of cHL is increasingly better understood (70), the roles of the EBV miRNAs have only just begun to be explored (Figure 2).
Epstein-Barr virus nuclear antigen-1 (EBNA1)
EBNA1 is essential for the maintenance of EBV episomes in infected cells, as it is a key viral replication factor and is responsible for tethering viral genomes to the chromosomes of the host cell; loss of EBNA1 expression therefore results in the loss of EBV genomes during cell division (71). EBNA1 is also a transcription factor that can bind to viral and cellular promoters (72-74). EBNA1 inhibits TGFβ signalling, in part by modulating the levels of SMAD2. EBNA1 also decreases expression of the TGFβ target gene, PTPRK, which in turn promotes the growth and survival of HRS cells (75,76).
Latent membrane protein-1
LMP1 is a constitutively active homologue of CD40 that has been shown to activate NF-κB, JAK/STAT, AP-1 and phosphatidylinositol-3 kinase (PI3K)/AKT signalling (77-79). While LMP1 might be responsible for activating these pathways in EBV-positive cHL, mutations in components of these signalling pathways appear to be necessary when the virus is absent. For example, TNFAIP3 mutations are more commonly seen in EBV-negative cHL, suggesting that mutational inactivation of TNFAIP3 and EBV infection are alternative pathways both leading to deregulated NF-κB signalling (19). The requirement for more genetic changes in EBV-negative cHL is supported by other studies which show that EBV-positive cHL has significantly fewer chromosome abnormalities than EBV-negative cHL (80-82). LMP1 also induces many of the features of the aberrant transcriptional programme characteristic of HRS cells, including the down-regulation of BCR signalling components, and the increased expression of anti-apoptotic genes such as BCL2 and BFL-1 (83,84). The FLICE-inhibitory protein (c-FLIP), a negative regulator of Fas induced apoptosis (85), is also an LMP1 target and could be important in the rescue of pre-apoptotic GC B cells early in cHL pathogenesis (86). LMP1 also increases the expression of Programmed death ligand-1 (PD-L1) in B cells, which might account at least in part for the sensitivity of cHL to immune checkpoint inhbitors targeting the PD-L1/PD-1 axis, although definitive studies assessing the sensitivity of EBV-positive versus EBV-negative cHL to immune checkpoint blockade have not been done (87).
Latent membrane protein-2A (LMP2A)
EBV appears to be crucial for the survival of HRS progenitors harbouring so called ‘crippling’ mutations in immunoglobulin genes; these mutations are found almost exclusively in EBV positive cases (88). EBV efficiently immortalises BCR-negative GC B cells in vitro; an effect that is dependent upon LMP2A which provides a BCR-like signalling function (61,89). LMP2A also contributes to the transcriptional programme of cHL, for example by reducing the expression of numerous B cell transcription factors, including EBF1 and E2A (90-93).
EBV-encoded miRNA
LCL express a subset of BART miRNA (including approximately half of Cluster 2) as well as 3 of the 4 BHRF1 miRNAs, that are subsequently turned off in GC B cells and memory B cells. In parallel, EBV-infected GC B cells and memory B cells up-regulate the remaining BART miRNAs by 5–10 fold (94). In EBV associated tumours expressing a latency II phenotype, including cHL, the latency III associated BART but not BHRF1 miRNAs are up-regulated (95). Among these, miR-BART2-5p is expressed in EBV-positive cHL and can act as an anti-sense miRNA to the EBV DNA polymerase BALF5 to inhibit virus replication (96). miR-BART2-5p also decreases BCR-mediated NF-kappa-B activation (97). miR-BART13-3p is one of the most highly expressed viral miRNA in cHL, and can be released into the circulation via exosomes (98). miR-BARTs in exosomes derived from EBV-positive cells have been shown to induce changes in the phenotype of macrophages, including the increased production of cytokines, such TNF-α, and IL-10 (99).
EBV also influences host miRNA expression in cHL. For example, Navarro et al. observed a subset of 10 host miRNAs whose expression was influenced by the presence of EBV. Among these, miR-96, miR-128a, miR-128b were selectively down regulated in EBV-positive cHL. The authors also reported a distinctive signature of 25 miRNAs that were differentially expressed between cHL and reactive lymph nodes. Interestingly, the dysregulation of miR-21 that favours cell survival by indirectly up-regulating anti-apoptotic genes suggests an important role for cellular miRNAs in the biology of cHL (100).
Co-expression of latent genes
A drawback of many of the studies investigating the function of individual virus genes has been the reliance on single gene over-expression or knockdown/knockout experiments; an approach that cannot take account of the effects of the co-expression of virus genes. This could be important, particularly because some studies have shown that some of the latent genes, for example, LMP1 and LMP2A have partially overlapping, as well as counteracting, transcriptional programmes (101). Moreover, it has been shown that when expressed in the B cells of transgenic mice, LMP1 is transforming, but not when LMP2A is co-expressed (102). In another study it was shown that LMP1 and LMP2A co-expression in mouse B cells led to tumour development, but only if the animals were immunosuppressed, suggesting that a background of immune impairment is important (103). Studies exploring the co-expression of virus genes in cHL at single cell resolution are required.
EBV and the tumour microenvironment of cHL
The TME is a defining feature of cHL and there is some evidence that EBV is at least partly responsible for reshaping this TME through expression of the latent genes, and also potentially through limited induction of the lytic cycle. For example, LMP1 an increase production of a diverse array of different chemokines and cytokines (104,105). EBNA1 can also influence the transcription of genes encoding key soluble factors, for example the chemokine CCL20 which is important in the chemo-attraction of regulatory T cells (106). Conversely, the cHL TME of cHL could also modulate viral gene expression in tumour cells. A good example here is the expression of LMP1 which can be regulated by different cytokines (107,108). The TME could also modify EBV’s oncogenic functions. For example, LMP1 can increase expression of discoidin domain receptor 1 (DDR1), a collagen receptor leading to the increased survival of lymphoma cells (109).
The function of EBV-specific T cells in the TME of cHL might be compromised by a variety of immune evasion mechanisms. Thus, loss of HLA expression is frequently observed in HRS cells, but is more commonly found in EBV-negative cHL (110-112); in some cases this is caused by inactivating mutations in the beta-2-microglobulin gene (113). EBV-positive cases of cHL usually express normal or sometimes higher levels of HLA expression, and contain more activated CTLs than EBV-negative cases (110-112,114). EBV-positive cHL also contains more NK cells (115). These data suggest that EBV utilises other mechanisms to evade anti-tumour immune responses in cHL, including the induction of immune checkpoint molecules, such as PD-L1 (87).
Difficulties in the diagnosis of EBV-positive cHL
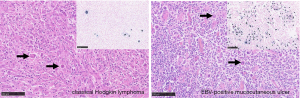
The diagnosis of cHL is dependent upon morphological evaluation by a haematopathologist who can recognize the typical HRS cell morphology and the characteristic cellular infiltrate supported by additional immunohistochemistry stains for example for recognition of CD30-positive HRS cells. However, a number of EBV-associated lymphoproliferative disorders (LPD) can harbour EBV-positive HRS-like B-cells, morphologically mimicking EBV-positive cHL. Many of these LPD show distinct clinical behaviour and require radically different management approaches from cHL, potentially leading to over- or undertreatment if not recognised by the pathologist. Whilst cHL is typically a nodal disease, extranodal extension can be seen in late stage disease (116). Furthermore, extremely rare manifestations of primary extra-nodal cHL have been described (117). Similarly, many of the mimics of EBV-positive cHL can present as nodal or extranodal disease and thus have to be considered in the differential diagnosis.
EBV-positive diffuse large B-cell lymphoma (DLBCL), NOS, is systemic lymphoma typically presenting with prominent extranodal involvement in the elderly, although younger patients can also be affected (118). EBV-positive DLBCL, NOS, can show large pleomorphic HRS-like lesional B-cells which can associated with a prominent inflammatory milieu. On immunophenotyping, CD30 expression is usually seen in the lesional cells and can co-exist with CD15 expression in 68% of cases (119). Expression of CD20 is an important marker in distinguishing EBV+ DLBCL, NOS, from EBV+ cHL. However, CD20 can show variable expression in EBV+ DLBCL, NOS, as over 50% of lesional cells are typically positive.
EBV-positive mucocutaneous ulcer (EBVMCU) is a localised ulcerating EBV-positive LPD affecting the skin or mucous membranes (120) (Figure 3). EBVMCU are usually solitary but multi-focal manifestations have been described (121). The lesions are typically associated with immunosuppression or in older patients, are assumed to be consequence of immunosenescence (120). EBVMCU is an indolent disease and usually resolves upon once the source of immunosuppression is removed. Rituximab has been used with some success in refractory cases.
Some lymphomas, not in themselves associated with EBV, can show EBV-positive HRS-like B-cells in the background. The archetypal example is angioimmunoblastic lymphoma (AIL), a T-cell lymphoma with a T-follicular helper(Tfh) cell phenotype (118). Up to 95% of AIL can show EBV+ B-cell blasts in the background. In some cases, the B-cell component in the background of AIL can progress to EBV+ DLBCL or EBV+ cHL (122). Other lymphomas accompanied by EBV+ B-cell blasts have also been described in the background of other EBV-negative lymphomas including chronic lymphocytic leukaemia and mantle zone lymphoma (123,124).
EBV-positive manifestations of lymphomas, traditionally considered not to be associated with EBV, have now been described, thus broadening the range of potential differential diagnoses. Rare examples of EBV-positive nodular lymphocyte predominant Hodgkin lymphoma, primary mediastinal B-cell lymphoma and EBV-positive mediastinal grey-zone lymphoma (PMGZL) (intermediate features between primary mediastinal lymphoma and Hodgkin lymphoma) have been described (125,126). The diagnosis of mediastinal grey-zone lymphoma is further complicated, as both cHL and PMGZL can occur in the same vicinity (composite lymphoma), or develop sequentially, where EBV status has been reported to change from the initial to subsequent biopsy. The diagnosis of these entities necessitates the presence of all the typical diagnostic features of the EBV-negative variant to be present in order to make the diagnosis. Nonetheless, the recognition of these entities can make distinction between the various EBV-positive LPD extremely difficult.
Concluding remarks
A subset of cHL patients harbor EBV in their tumour cells. Although the contribution of EBV to the pathogenesis of cHL has been debated, we argue that differences in the epidemiology, genetics, and biology of EBV-positive compared with EBV-negative cHL strongly suggest a key role for the virus. Notwithstanding debate over EBV’s exact contribution to the oncogenic process in cHL, there is undoubtedly an opportunity to exploit the presence of EBV in tumours for patient benefit. In some LPD and in other EBV-associated tumours such as nasopharyngeal carcinoma, EBV detection in blood and other tissues can be a useful biomarker. However, as yet routine blood testing for EBV is not employed in the management of the majority of EBV-associated malignancies, including cHL. Moreover, the development of new approaches to specifically target EBV, for example, by reactivating the virus into the lytic cycle (and thereby killing the infected cell, or making it susceptible to drugs such as ganciclovir) (127-129), gene therapy (130), EBNA1 inhibitors (131,132), and therapeutic vaccination/T cell therapies (133,134) are provide exciting new opportunities to improve outcomes for patients.
Acknowledgments
We are grateful to the Children’s Cancer and Leukaemia Group (CCLG; PM, KV), Blood Cancer UK (PM, KV), the Medical Research Council, UK (MP) and an EU Marie Curie Sklodowska Fellowship (LM).
Funding: The work was supported in part by a European Regional Development Fund Project (ENOCH: CZ.02.1.01/0.0/0.0/16_019/0000868).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Christopher P. Fox, Claire Shannon-Lowe) for the series “Lymphoma and Viruses” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-21-8). The series “Lymphoma and Viruses” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: IARC Press; 2008.
- Küppers R, Rajewsky K, Zhao M, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A 1994;91:10962-6. [Crossref] [PubMed]
- Kanzler H, Kuppers R, Hansmann ML, et al. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996;184:1495-505. [Crossref] [PubMed]
- Vockerodt M, Soares M, Kanzler H, et al. Detection of clonal Hodgkin and Reed-Sternberg cells with identical somatically mutated and rearranged VH genes in different biopsies in relapsed Hodgkin's disease. Blood 1998;92:2899-907. [Crossref] [PubMed]
- Marafioti T, Hummel M, Foss HD, et al. Hodgkin and reed-sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000;95:1443-50. [Crossref] [PubMed]
- Bargou RC, Leng C, Krappmann D, et al. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood 1996;87:4340-7. [Crossref] [PubMed]
- Izban KF, Ergin M, Huang Q, et al. Characterization of NF-kappaB expression in Hodgkin's disease: inhibition of constitutively expressed NF-kappaB results in spontaneous caspase-independent apoptosis in Hodgkin and Reed-Sternberg cells. Mod Pathol 2001;14:297-310. [Crossref] [PubMed]
- Bargou RC, Emmerich F, Krappmann D, et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J Clin Invest 1997;100:2961-9. [Crossref] [PubMed]
- Carbone A, Gloghini A, Gruss HJ, et al. CD40 ligand is constitutively expressed in a subset of T cell lymphomas and on the microenvironmental reactive T cells of follicular lymphomas and Hodgkin's disease. Am J Pathol 1995;147:912-22. [PubMed]
- Pinto A, Aldinucci D, Gloghini A, et al. The role of eosinophils in the pathobiology of Hodgkin's disease. Ann Oncol 1997;8:89-96. [Crossref] [PubMed]
- Barth TF, Martin-Subero JI, Joos S, et al. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood 2003;101:3681-6. [Crossref] [PubMed]
- Joos S, Granzow M, Holtgreve-Grez H, et al. Hodgkin's lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer 2003;103:489-95. [Crossref] [PubMed]
- Martín-Subero JI, Gesk S, Harder L, et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 2002;99:1474-7. [Crossref] [PubMed]
- Cabannes E, Khan G, Aillet F, et al. Mutations in the IkBa gene in Hodgkin's disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 1999;18:3063-70. [Crossref] [PubMed]
- Emmerich F, Meiser M, Hummel M, et al. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 1999;94:3129-34. [Crossref] [PubMed]
- Jungnickel B, Staratschek-Jox A, Brauninger A, et al. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin's lymphoma. J Exp Med 2000;191:395-402. [Crossref] [PubMed]
- Emmerich F, Theurich S, Hummel M, et al. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J Pathol 2003;201:413-20. [Crossref] [PubMed]
- Lake A, Shield LA, Cordano P, et al. Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int J Cancer 2009;125:1334-42. [Crossref] [PubMed]
- Schmitz R, Hansmann ML, Bohle V, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 2009;206:981-9. [Crossref] [PubMed]
- Hinz M, Lemke P, Anagnostopoulos I, et al. Nuclear factor kappaB-dependent gene expression profiling of Hodgkin's disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity. J Exp Med 2002;196:605-17. [Crossref] [PubMed]
- Skinnider BF, Elia AJ, Gascoyne RD, et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002;99:618-26. [Crossref] [PubMed]
- Kube D, Holtick U, Vockerodt M, et al. STAT3 is constitutively activated in Hodgkin cell lines. Blood 2001;98:762-70. [Crossref] [PubMed]
- Joos S, Kupper M, Ohl S, et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000;60:549-52. [PubMed]
- Weniger MA, Melzner I, Menz CK, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006;25:2679-84. [Crossref] [PubMed]
- Gunawardana J, Chan FC, Telenius A, et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet 2014;46:329-35. [Crossref] [PubMed]
- Levine PH AD, Berard CW, Carbone PP, Waggoner DE, Malan L. Elevated antibody titers to Epstein-Barr virus in Hodgkin's disease. Cancer 1971;27:416-21. [Crossref] [PubMed]
- Mueller N, Evans A, Harris NL, et al. Hodgkin's disease and Epstein-Barr virus. Altered antibody pattern before diagnosis. N Engl J Med 1989;320:689-95. [Crossref] [PubMed]
- Weiss LM, Strickler JG, Warnke RA, et al. Epstein-Barr viral DNA in tissues of Hodgkin's disease. Am J Pathol 1987;129:86-91. [PubMed]
- Poppema S, van Imhoff G, Torensma R, et al. Lymphadenopathy morphologically consistent with Hodgkin's disease associated with Epstein-Barr virus infection. Am J Clin Pathol 1985;84:385-90. [Crossref] [PubMed]
- Anagnostopoulos I, Herbst H, Niedobitek G, et al. Demonstration of monoclonal EBV genomes in Hodgkin's disease and Ki-1-positive anaplastic large cell lymphoma by combined Southern blot and in situ hybridization. Blood 1989;74:810-6. [Crossref] [PubMed]
- Weiss LM, Movahed LA, Warnke RA, et al. Detection of Epstein–Barr Viral Genomes in Reed–Sternberg Cells of Hodgkin's Disease. N Engl J Med 1989;320:502-6. [Crossref] [PubMed]
- Wu T-C, Mann RB, Charache P, et al. Detection of EBV gene expression in Reed-Sternberg cells of Hodgkin's disease. Int J Cancer 1990;46:801-4. [Crossref] [PubMed]
- Coates PJ, Slavin G, D'Ardenne AJ. Persistence of Epstein-Barr virus in Reed-Sternberg cells throughout the course of Hodgkin's disease. J Pathol 1991;164:291-7. [Crossref] [PubMed]
- Glaser SL, Lin RJ, Stewart SL, et al. Epstein-Barr virus-associated Hodgkin's disease: epidemiologic characteristics in international data. Int J Cancer 1997;70:375-82. [Crossref] [PubMed]
- Glaser SL, Jarrett RF. The epidemiology of Hodgkin's disease. Baillieres Clin Haematol 1996;9:401-16. [Crossref] [PubMed]
- Armstrong AA, Alexander FE, Cartwright R, et al. Epstein-Barr virus and Hodgkin's disease: further evidence for the three disease hypothesis. Leukemia 1998;12:1272-6. [Crossref] [PubMed]
- Jarrett RF, Gallagher A, Jones DB, et al. Detection of Epstein-Barr virus genomes in Hodgkin's disease: relation to age. J Clin Pathol 1991;44:844-8. [Crossref] [PubMed]
- Chang KL, Albujar PF, Chen YY, et al. High prevalence of Epstein-Barr virus in the Reed-Sternberg cells of Hodgkin's disease occurring in Peru. Blood 1993;81:496-501. [Crossref] [PubMed]
- Weinreb M, Day PJ, Niggli F, et al. The consistent association between Epstein-Barr virus and Hodgkin's disease in children in Kenya. Blood 1996;87:3828-36. [Crossref] [PubMed]
- Flavell KJ, Biddulph JP, Powell JE, et al. South Asian ethnicity and material deprivation increase the risk of Epstein-Barr virus infection in childhood Hodgkin's disease. Br J Cancer 2001;85:350-6. [Crossref] [PubMed]
- Biggar RJ, Jaffe ES, Goedert JJ, et al. Hodgkin lymphoma and immunodeficiency in persons with HIV/AIDS. Blood 2006;108:3786-91. [Crossref] [PubMed]
- Glaser SL, Clarke CA, Gulley ML, et al. Population-based patterns of human immunodeficiency virus-related Hodgkin lymphoma in the Greater San Francisco Bay Area, 1988-1998. Cancer 2003;98:300-9. [Crossref] [PubMed]
- Crump C, Sundquist K, Sieh W, et al. Perinatal and family risk factors for Hodgkin lymphoma in childhood through young adulthood. Am J Epidemiol 2012;176:1147-58. [Crossref] [PubMed]
- Crump C, Sundquist K, Sieh W, et al. Perinatal and family risk factors for non-Hodgkin lymphoma in early life: a Swedish national cohort study. J Natl Cancer Inst 2012;104:923-30. [Crossref] [PubMed]
- Mack TM, Cozen W, Shibata DK, et al. Concordance for Hodgkin's disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. N Engl J Med 1995;332:413-8. [Crossref] [PubMed]
- Kushekhar K, van den Berg A, Nolte I, et al. Genetic associations in classical hodgkin lymphoma: a systematic review and insights into susceptibility mechanisms. Cancer Epidemiol Biomarkers Prev 2014;23:2737-47. [Crossref] [PubMed]
- Hjalgrim H, Rostgaard K, Johnson PC, et al. HLA-A alleles and infectious mononucleosis suggest a critical role for cytotoxic T-cell response in EBV-related Hodgkin lymphoma. Proc Natl Acad Sci U S A 2010;107:6400-5. [Crossref] [PubMed]
- Niens M, Jarrett RF, Hepkema B, et al. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood 2007;110:3310-5. [Crossref] [PubMed]
- Niens M, van den Berg A, Diepstra A, et al. The human leukocyte antigen class I region is associated with EBV-positive Hodgkin's lymphoma: HLA-A and HLA complex group 9 are putative candidate genes. Cancer Epidemiol Biomarkers Prev 2006;15:2280-4. [Crossref] [PubMed]
- Farrell K, Jarrett RF. The molecular pathogenesis of Hodgkin lymphoma. Histopathology 2011;58:15-25. [Crossref] [PubMed]
- Connelly RR, Christine BW. A cohort study of cancer following infectious mononucleosis. Cancer Res 1974;34:1172-8. [PubMed]
- Rosdahl N, Larsen SO, Clemmesen J. Hodgkin's disease in patients with previous infectious mononucleosis: 30 years' experience. Br Med J 1974;2:253-6. [Crossref] [PubMed]
- Hjalgrim H, Smedby KE, Rostgaard K, et al. Infectious mononucleosis, childhood social environment, and risk of Hodgkin lymphoma. Cancer Res 2007;67:2382-8. [Crossref] [PubMed]
- Rickinson AB, Rowe M, Hart IJ, et al. T-cell-mediated regression of "spontaneous" and of Epstein-Barr virus-induced B-cell transformation in vitro: studies with cyclosporin A. Cell Immunol 1984;87:646-58. [Crossref] [PubMed]
- Kerr BM, Lear AL, Rowe M, et al. Three transcriptionally distinct forms of epstein-barr virus latency in somatic cell hybrids: Cell phenotype dependence of virus promoter usage. Virology 1992;187:189-201. [Crossref] [PubMed]
- Pfeffer S. Identification of virus-encoded MicroRNAs. Science 2004;304:734-6. [Crossref] [PubMed]
- Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 2016;16:789-802. [Crossref] [PubMed]
- Babcock GJ, Decker LL, Volk M, et al. EBV persistence in memory B cells in vivo. Immunity 1998;9:395-404. [Crossref] [PubMed]
- Babcock GJ, Hochberg D, Thorley-Lawson DA. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 2000;13:497-506. [Crossref] [PubMed]
- Gires O, Zimber-Strobl U, Gonnella R, et al. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J 1997;16:6131-40. [Crossref] [PubMed]
- Caldwell RG, Wilson JB, Anderson SJ, et al. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998;9:405-11. [Crossref] [PubMed]
- Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the Replicative cycle of Epstein-Barr virus in vivo. J Virol 2005;79:1296-307. [Crossref] [PubMed]
- Kurth J, Hansmann ML, Rajewsky K, et al. Epstein-Barr virus-infected B cells expanding in germinal centers of infectious mononucleosis patients do not participate in the germinal center reaction. Proc Natl Acad Sci U S A 2003;100:4730-5. [Crossref] [PubMed]
- Kurth J, Spieker T, Wustrow J, et al. EBV-Infected B cells in infectious Mononucleosis. Immunity 2000;13:485-95. [Crossref] [PubMed]
- Mohamed G, Vrzalikova K, Cader FZ, et al. Epstein-Barr virus, the germinal centre and the development of Hodgkin's lymphoma. J Gen Virol 2014;95:1861-9. [Crossref] [PubMed]
- Chaganti S, Ma CS, Bell AI, et al. Epstein-Barr virus persistence in the absence of conventional memory B cells: IgM+IgD+CD27+ B cells harbor the virus in X-linked lymphoproliferative disease patients. Blood 2008;112:672-9. [Crossref] [PubMed]
- Agematsu K, Nagumo H, Shinozaki K, et al. Absence of IgD-CD27(+) memory B cell population in X-linked hyper-IgM syndrome. J Clin Invest 1998;102:853-60. [Crossref] [PubMed]
- Ma CS, Pittaluga S, Avery DT, et al. Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest 2006;116:322-33. [Crossref] [PubMed]
- Heath E, Begue-Pastor N, Chaganti S, et al. Epstein-Barr virus infection of naive B cells in vitro frequently selects clones with mutated immunoglobulin genotypes: implications for virus biology. PLoS Pathog 2012;8:e1002697 [Crossref] [PubMed]
- Murray PG, Young LS. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood 2019;134:591-6. [Crossref] [PubMed]
- Westhoff Smith D, Sugden B. Potential cellular functions of Epstein-Barr Nuclear Antigen 1 (EBNA1) of Epstein-Barr Virus. Viruses 2013;5:226-40. [Crossref] [PubMed]
- Frappier L. Contributions of Epstein-Barr nuclear antigen 1 (EBNA1) to cell immortalization and survival. Viruses 2012;4:1537-47. [Crossref] [PubMed]
- Frappier L. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr Opin Virol 2012;2:733-9. [Crossref] [PubMed]
- Frappier L. The Epstein-Barr Virus EBNA1 Protein. Scientifica (Cairo) 2012;2012:438204 [Crossref] [PubMed]
- Flavell JR, Baumforth KR, Wood VH, et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood 2008;111:292-301. [Crossref] [PubMed]
- Wood VH, O'Neil JD, Wei W, et al. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene 2007;26:4135-47. [Crossref] [PubMed]
- Floettmann JE, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerisation for NF-kappaB activation. Oncogene 1997;15:1851-8. [Crossref] [PubMed]
- Shair KH, Bendt KM, Edwards RH, et al. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog 2007;3:e166 [Crossref] [PubMed]
- Zhang L, Hong K, Zhang J, et al. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology 2004;323:141-52. [Crossref] [PubMed]
- Wienand K, Chapuy B, Stewart C, et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv 2019;3:4065-80. [Crossref] [PubMed]
- Montgomery ND. Karyotypic abnormalities associated with Epstein-Barr virus status in classical Hodgkin lymphoma. Cancer Genet 2016;209:408-16. [Crossref] [PubMed]
- Tiacci E, Ladewig E, Schiavoni G, et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018;131:2454-65. [Crossref] [PubMed]
- Henderson S, Rowe M, Gregory C, et al. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 1991;65:1107-15. [Crossref] [PubMed]
- Vockerodt M, Morgan SL, Kuo M, et al. The Epstein-Barr virus oncoprotein, latent membrane protein-1, reprograms germinal centre B cells towards a Hodgkin's Reed-Sternberg-like phenotype. The Journal of Pathology 2008;216:83-92. [Crossref] [PubMed]
- Cahir-McFarland ED, Carter K, Rosenwald A, et al. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1 - Expressing or Epstein-Barr virus latency III-infected cells. Journal of Virology 2004;78:4108-19. [Crossref] [PubMed]
- Dutton A, O'Neil JD, Milner AE, et al. Expression of the cellular FLICE-inhibitory protein (c-FLIP) protects Hodgkin's lymphoma cells from autonomous Fas-mediated death. Proc Natl Acad Sci U S A 2004;101:6611-6. [Crossref] [PubMed]
- Green MR, Rodig S, Juszczynski P, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res 2012;18:1611-8. [Crossref] [PubMed]
- Bräuninger A, Schmitz R, Bechtel D, et al. Molecular biology of Hodgkin's and Reed/Sternberg cells in Hodgkin's lymphoma. Int J Cancer 2006;118:1853-61. [Crossref] [PubMed]
- Merchant M, Swart R, Katzman RB, et al. The effects of the Epstein-Barr virus latent membrane protein 2A on B cell function. Int Rev Immunol 2001;20:805-35. [Crossref] [PubMed]
- Portis T, Dyck P, Longnecker R. Epstein-Barr Virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood 2003;102:4166-78. [Crossref] [PubMed]
- Portis T, Longnecker R. Epstein-Barr virus (EBV) LMP2A alters normal transcriptional regulation following B-cell receptor activation. Virology 2004;318:524-33. [Crossref] [PubMed]
- Portis T, Longnecker R. Epstein-Barr virus LMP2A interferes with global transcription factor regulation when expressed during B-lymphocyte development. J Virol 2003;77:105-14. [Crossref] [PubMed]
- Vockerodt M, Wei W, Nagy E, et al. Suppression of the LMP2A target gene, EGR-1, protects Hodgkin's lymphoma cells from entry to the EBV lytic cycle. J Pathol 2013;230:399-409. [Crossref] [PubMed]
- Qiu J, Smith P, Leahy L, et al. The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog 2015;11:e1004561 [Crossref] [PubMed]
- Qiu J, Cosmopoulos K, Pegtel M, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog 2011;7:e1002193 [Crossref] [PubMed]
- Barth S, Pfuhl T, Mamiani A, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res 2008;36:666-75. [Crossref] [PubMed]
- Chen Y, Fachko D, Ivanov NS, et al. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog 2019;15:e1007535 [Crossref] [PubMed]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010;107:6328-33. [Crossref] [PubMed]
- Higuchi H, Yamakawa N, Imadome KI, et al. Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood 2018;131:2552-67. [Crossref] [PubMed]
- Navarro A, Gaya A, Martinez A, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood 2008;111:2825-32. [Crossref] [PubMed]
- Vrzalikova K, Ibrahim M, Nagy E, et al. Co-Expression of the Epstein-Barr Virus-Encoded Latent Membrane Proteins and the Pathogenesis of Classic Hodgkin Lymphoma. Cancers (Basel) 2018;10:285. [Crossref] [PubMed]
- Vrazo AC, Chauchard M, Raab-Traub N, et al. Epstein-Barr virus LMP2A reduces hyperactivation induced by LMP1 to restore normal B cell phenotype in transgenic mice. PLoS Pathog 2012;8:e1002662 [Crossref] [PubMed]
- Wirtz T, Weber T, Kracker S, et al. Mouse model for acute Epstein-Barr virus infection. Proc Natl Acad Sci U S A 2016;113:13821-6. [Crossref] [PubMed]
- Vockerodt M, Pinkert D, Smola-Hess S, et al. The Epstein-Barr virus oncoprotein latent membrane protein 1 induces expression of the chemokine IP-10: importance of mRNA half-life regulation. Int J Cancer 2005;114:598-605. [Crossref] [PubMed]
- Nakagomi H, Dolcetti R, Bejarano MT, et al. The Epstein-Barr virus latent membrane protein-1 (LMP1) induces interleukin-10 production in Burkitt lymphoma lines. Int J Cancer 1994;57:240-4. [Crossref] [PubMed]
- Baumforth KR, Birgersdotter A, Reynolds GM, et al. Expression of the Epstein-Barr virus-encoded Epstein-Barr virus nuclear antigen 1 in Hodgkin's lymphoma cells mediates Up-regulation of CCL20 and the migration of regulatory T cells. Am J Pathol 2008;173:195-204. [Crossref] [PubMed]
- Kis LL, Gerasimcik N, Salamon D, et al. STAT6 signaling pathway activated by the cytokines IL-4 and IL-13 induces expression of the Epstein-Barr virus-encoded protein LMP-1 in absence of EBNA-2: implications for the type II EBV latent gene expression in Hodgkin lymphoma. Blood 2011;117:165-74. [Crossref] [PubMed]
- Kis LL, Takahara M, Nagy N, et al. Cytokine mediated induction of the major Epstein-Barr virus (EBV)-encoded transforming protein, LMP-1. Immunol Lett 2006;104:83-8. [Crossref] [PubMed]
- Cader FZ, Vockerodt M, Bose S, et al. The EBV oncogene LMP1 protects lymphoma cells from cell death through the collagen-mediated activation of DDR1. Blood 2013;122:4237-45. [Crossref] [PubMed]
- Lee SP, Constandinou CM, Thomas WA, et al. Antigen presenting phenotype of Hodgkin Reed-Sternberg cells: analysis of the HLA class I processing pathway and the effects of interleukin-10 on epstein-barr virus-specific cytotoxic T-cell recognition. Blood 1998;92:1020-30. [Crossref] [PubMed]
- Murray PG, Constandinou CM, Crocker J, et al. Analysis of major histocompatibility complex class I, TAP expression, and LMP2 epitope sequence in Epstein-Barr virus-positive Hodgkin's disease. Blood 1998;92:2477-83. [Crossref] [PubMed]
- Oudejans JJ, Jiwa NM, Kummer JA, et al. Analysis of major histocompatibility complex class I expression on Reed-Sternberg cells in relation to the cytotoxic T-cell response in Epstein-Barr virus-positive and -negative Hodgkin's disease. Blood 1996;87:3844-51. [Crossref] [PubMed]
- Reichel J, Chadburn A, Rubinstein PG, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015;125:1061-72. [Crossref] [PubMed]
- Oudejans JJ, Jiwa NM, Kummer JA, et al. Activated cytotoxic T cells as prognostic marker in Hodgkin's disease. Blood 1997;89:1376-82. [Crossref] [PubMed]
- Wu R, Sattarzadeh A, Rutgers B, et al. The microenvironment of classical Hodgkin lymphoma: heterogeneity by Epstein-Barr virus presence and location within the tumor. Blood Cancer J 2018;8:e622 [Crossref] [PubMed]
- Cheson BD, Fisher RI, Barrington SF, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 2014;32:3059-68. [Crossref] [PubMed]
- Yang M, Ping L, Liu W, et al. Clinical characteristics and prognostic factors of primary extranodal classical Hodgkin lymphoma: a retrospective study. Hematology 2019;24:413-9. [Crossref] [PubMed]
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391-405. [Crossref] [PubMed]
- Dojcinov SD, Venkataraman G, Pittaluga S, et al. Age-related EBV-associated lymphoproliferative disorders in the Western population: a spectrum of reactive lymphoid hyperplasia and lymphoma. Blood 2011;117:4726-35. [Crossref] [PubMed]
- Dojcinov SD, Venkataraman G, Raffeld M, et al. EBV positive mucocutaneous ulcer--a study of 26 cases associated with various sources of immunosuppression. Am J Surg Pathol 2010;34:405-17. [Crossref] [PubMed]
- Pugh MR, Leopold GD, Morgan M, et al. Epstein-Barr Virus-Positive Mucocutaneous Ulcers Complicate Colitis Caused by Immune Checkpoint Regulator Therapy and Associate With Colon Perforation. Clin Gastroenterol Hepatol 2020;18:1785-95.e3. [Crossref] [PubMed]
- Attygalle AD, Kyriakou C, Dupuis J, et al. Histologic evolution of angioimmunoblastic T-cell lymphoma in consecutive biopsies: clinical correlation and insights into natural history and disease progression. Am J Surg Pathol 2007;31:1077-88. [Crossref] [PubMed]
- Kanzler H, Kuppers R, Helmes S, et al. Hodgkin and Reed-Sternberg-like cells in B-cell chronic lymphocytic leukemia represent the outgrowth of single germinal-center B-cell-derived clones: potential precursors of Hodgkin and Reed-Sternberg cells in Hodgkin's disease. Blood 2000;95:1023-31. [Crossref] [PubMed]
- Kanai R, Miyagawa-Hayashino A, Shishido-Hara Y, et al. Mantle cell lymphoma with EBV-positive Hodgkin and Reed-Sternberg-like cells in a patient after autologous PBSCT: Phenotypically distinct but genetically related tumors. Pathol Int 2021;71:96-101. [Crossref] [PubMed]
- Huppmann AR, Nicolae A, Slack GW, et al. EBV may be expressed in the LP cells of nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) in both children and adults. Am J Surg Pathol 2014;38:316-24. [Crossref] [PubMed]
- Elsayed AA, Satou A, Eladl AE, et al. Grey zone lymphoma with features intermediate between diffuse large B-cell lymphoma and classical Hodgkin lymphoma: a clinicopathological study of 14 Epstein-Barr virus-positive cases. Histopathology 2017;70:579-94. [Crossref] [PubMed]
- Kerr JR. Epstein-Barr virus (EBV) reactivation and therapeutic inhibitors. J Clin Pathol 2019;72:651-8. [Crossref] [PubMed]
- Tikhmyanova N, Paparoidamis N, Romero-Masters J, et al. Development of a novel inducer for EBV lytic therapy. Bioorg Med Chem Lett 2019;29:2259-64. [Crossref] [PubMed]
- Ramos JC, Sparano JA, Rudek MA, et al. Safety and Preliminary Efficacy of Vorinostat With R-EPOCH in High-risk HIV-associated Non-Hodgkin's Lymphoma (AMC-075). Clin Lymphoma Myeloma Leuk 2018;18:180-90.e2. [Crossref] [PubMed]
- Ricciardelli I, Blundell MP, Brewin J, et al. Towards gene therapy for EBV-associated posttransplant lymphoma with genetically modified EBV-specific cytotoxic T cells. Blood 2014;124:2514-22. [Crossref] [PubMed]
- Messick TE, Smith GR, Soldan SS, et al. Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci Transl Med 2019;11:eaau5612 [Crossref] [PubMed]
- Jiang L, Xie C, Lung HL, et al. EBNA1-targeted inhibitors: Novel approaches for the treatment of Epstein-Barr virus-associated cancers. Theranostics 2018;8:5307-19. [Crossref] [PubMed]
- Taylor GS, Jia H, Harrington K, et al. A recombinant modified vaccinia ankara vaccine encoding Epstein-Barr Virus (EBV) target antigens: a phase I trial in UK patients with EBV-positive cancer. Clin Cancer Res 2014;20:5009-22. [Crossref] [PubMed]
- Rühl J, Citterio C, Engelmann C, et al. Heterologous prime-boost vaccination protects against EBV antigen-expressing lymphomas. J Clin Invest 2019;129:2071-87. [Crossref] [PubMed]
Cite this article as: Vrzalikova K, Pugh M, Mundo L, Murray P. The contribution of ebv to the pathogenesis of classical hodgkin lymphoma. Ann Lymphoma 2021;5:30.