
Reframing Burkitt lymphoma: virology not epidemiology defines clinical variants
History of the discovery of Burkitt lymphoma (BL)
In 1958, Denis Burkitt described the cancer that is now known as BL (1). Although at first thought to be a sarcoma, BL was subsequently identified as a non-Hodgkin B cell lymphoma. The cancer typically presents extranodally and it was the unique presentation in the jaws of children that initially caught Burkitt’s attention. The finding that BL was restricted geographically, led to initial speculation that the tumor was caused by a mosquito borne virus (2,3). Denis Burkitt presented his observations of the tumor at Middlesex Hospital in London. In the audience was Tony Epstein who, intrigued by the idea of an infectious etiology of the cancer, initiated a collaboration with Burkitt. This ultimately led to the identification of the virus, Epstein-Barr virus (EBV) (4), the first virus shown to be associated with cancer in humans. Since these initial discoveries, research on the role of EBV and BL continues to contribute to our understanding of oncogenesis and the role of viruses in driving malignancy.
Epidemiology of BL
Three clinical variants of BL have been described based on the epidemiology of the cancer: endemic, sporadic and immunodeficiency-associated (ID). The most common, and the variant initially described by Burkitt, is the endemic form of BL. Endemic BL (eBL) is a pediatric cancer with a peak incidence between 6–8 years of age and a predominance in males (5,6). eBL is found in regions of the world where malaria transmission is year-round, predominantly sub-Saharan Africa and Papua New Guinea (6-9). eBL presents extranodally, frequently in the jaw in younger children and abdominally in older children (10). Recent studies indicate a changing pattern of presentation with less tumors occurring in the jaw than historically reported (11). EBV is detected in ~95% of cases (12,13).
In contrast to eBL, cases of sporadic BL (sBL) occur throughout the world. It is a rare malignancy and can occur in both children, young adults as well as the elderly (14,15). sBL represents ~50% of childhood lymphomas but less than 3% of all lymphomas in USA and Western Europe (16). The incidence of sBL in US children was reported to be 2.5 cases per million person-years (17). In contrast to eBL, EBV is detected in only 10–30% of sBL (18,19). In one study, 38% of pediatric sBL were found to be EBV-positive (20). sBL, like eBL, also occurs more frequently in males, typically presents within the abdominal region and is found in lymph nodes as well as extranodally (19).
A third subtype of BL emerged along with the HIV epidemic, HIV-associated BL (also called AIDS-related and more recently defined as ID-BL). HIV-associated BL occurs early in HIV infection and before CD4+ T cell numbers drop (21,22). The incidence of HIV-associated BL has remained stable in the US since the introduction of combination antiretroviral therapy for HIV (23). This form of BL has 20–40% of cases that are EBV positive (24). However, the frequency of EBV+ tumors has been reported to be as high as 71% in a case series from Brazil (25). There are also a few case reports describing the occurrence of BL in post-transplant immunodeficient patients but this cancer is not common in this setting (26-29).
A different epidemiologic pattern of BL is found in South America where EBV is detected in approximately half of the cases of all ages (25,30-32). Some studies provided comparison of EBV positivity in BL occurring in children <5 years to children 5–16 years (33,34). Only children less than 5 years of age are consistently EBV+. This is in sharp contrast with eBL where there is no age dependent segregation of EBV positivity in children (20). Interestingly, the prevalence of EBV in BL varies between different regions within Brazil with regions in the North showing a higher frequency of EBV+ BL (25).
The variable association of the BL epidemiologic subtypes with EBV presents a challenge to our understanding of the etiology of this malignancy. Some have argued that because EBV is not consistently detected in different BL subtypes, EBV plays a non-pathogenetic role (35,36). However, as will be reviewed below, an argument can be made for distinguishing BL based on the presence or absence of EBV, not on the epidemiology of the tumor (e.g., sporadic vs. endemic). This has important implications for understanding the etiopathogenesis of the different BL clinical subtypes.
Pathology of BL
Part of the challenge in determining whether the endemic and sporadic forms of BL are distinct entities is the consistent histologic description of these tumors regardless of their epidemiologic origins. Dennis Wright was the first to describe the so-called “starry sky” appearance that is readily observed in cytologic preparations (37). This feature is due to the presence of a high mitotic index, marked apoptosis, and presence of tingible body macrophages. According to the WHO classification, BL are composed of monomorphic CD19+CD20+ B cells with basophilic cytoplasm (19), commonly IgM+ (38,39) and a Ki-67 score of ≥95% (19). B cell lineage markers indicative of centroblasts are seen (e.g., CD10, BCL6, CD38bright) (38,40) and germinal center gene expression program are also found (41) consistent with BL derived from centroblasts. Gene expression profiling of the different BL subtypes identified a common transcriptional program that is distinct from other non-Hodgkin B-cell lymphomas such as diffuse large B-cell lymphoma and with a profile similar to a germinal center B-cell origin (41-43). This molecular definition of BL, while important for understanding etiopathogenesis and development of new therapeutic approaches, is not feasible for routine diagnostic purposes.
All BL express high levels of MYC and greater than 90% have the characteristic translocation of the MYC oncogene (8q24) onto the immunoglobulin heavy (IgH) (14q34) in the majority of cases (>85%) [reviewed in (44)]. Infrequently, MYC translocates to the immunoglobulin light chain κ (2p12) or λ (22q11) locus (45). The translocation of MYC near immunoglobulin regulatory regions results in constitutive expression of MYC. MYC transcription is driven from the translocated allele (44,46). The chromosomal break points in both MYC and IgH vary between sBL and eBL (20) which argues for a different etiologic driver. The MYC translocation resulting in constitutive MYC expression is thought to be a key pathogenetic event leading to the emergence of a malignant clone (47).
The role of EBV
Despite almost 60 years since the discovery of EBV in a BL tumor cell, there are still many unsolved questions as to how EBV contributes to BL oncogenesis. The viral genome is detected in the majority of eBL cases (12,19). Subsequent analysis of EBV genomes in the tumor cells showed that the viral genome was clonal indicative that EBV infection preceeded the malignant transformation event (48). In a seminal prospective study in Uganda in the 1970’s, antibodies to EBV viral capsid antigen (VCA) were increased significantly prior to the emergence of the tumor providing further evidence of EBV’s causal role (49). Loss of the EBV genome from BL cell lines results in apoptotic cell death (50,51). In addition, EBV encodes several latent proteins [Epstein-Barr nuclear antigen (EBNA)-1, -2, -3A, -3C and latent membrane protein (LMP)-1] that are essential for B-cell transformation [reviewed in (52)].
One of the paradoxes of determining EBV’s role in BL oncogenesis is the fact that only one EBV latent protein, EBNA-1, is consistently expressed in BL (53). Studies to examine the transforming capacity of EBNA-1 have yielded differing results. Wilson generated a transgenic mouse that expressed EBNA1 which resulted in B cell lymphomas (54). However, Kang et al., was not able to reproduce this result (55). It should be noted that different mouse strains and different constructs for generation of EBNA-1 transgenic mice were used in these two studies. Inhibition of EBNA-1 in BL cell lines results in apoptosis (50,55). Consistent with this, Holowaty showed that EBNA-1 binds to USP7 (also called HAUSP), a deubiquitinase. USP7 binds to p53 and mdm2, stabilizing the proteins. Binding of EBNA-1 to USP7 destabilizes p53 potentially abrogating apoptotic responses (56). Of note, USP7 is mutated in EBV+ BL (39).
In addition to EBNA-1, the RNA-pol III non-translated RNAs termed EBV-encoded small RNAs (EBER)-1 and EBER-2 are also consistently expressed in BL (57,58) as well as all latently infected cells. Because of their high level of expression, the EBERs are readily detected by in situ hybridization and thus useful for identifying EBV+ cells in biopsies. Their functional role in EBV+ BL remains controversial. Takada and colleagues used an EBV+ BL cell line, termed Akata, that over time lost the EBV episome. They were able to show that introduction of EBER-1 restored resistance to apoptosis in this line via induction of BCL-2 (59). While this is an important study, it also highlights the challenge with addressing requirements for EBV-encoded RNA or EBV latent proteins in malignant transformation by using cell lines which are typically derived from BL biopsies and thus, not representative of a pre-malignant state where EBV is most likely to contribute to oncogenesis.
EBV-encoded BART miRNAs are also detected in EBV+ BL (57,60-62). The presence of the BART miRNAs in EBV+ BL was associated with a different transcriptional program (63) pointing to a possible oncogenic role. Forced loss of the EBV episome followed by expression of BART miRNAs rescues the cells from apoptosis via inactivation of CASP3 (64). Further evidence of an important role for miRNAs in BL comes from a study by Mundo et al. (65) where they were able to detect EBV miRNA in EBER-negative BL cells.
Other EBV latent and lytic transcripts and/or proteins have been detected in BL (57,58,66) but detection of these other viral transcripts is sporadic, not in the majority of tumor cells and the potential role for expression of these additional EBV transcripts in oncogenesis is not clear. There is one study that shows that detection of the immediate early protein, BZLF1 (also called ZEBRA) correlated with improved response to treatment (67). A subset of BL was identified that have a deletion of EBNA-2 gene and but express EBNA-3C (68) suggesting that EBNA-2, a MYC antagonist, needs to be downregulated either through transcriptional repression or through deletion.
A common theme emerges from a number of studies suggests that a prime role for EBV is to disarm apoptotic triggers in B-cells such as the over-expression of MYC (69). The corollary idea is that following disarmament of apoptosis, the requirement for expression of any EBV gene is unnecessary (36,70).
For the EBV− BL subtype, a hit and run mechanism has been proposed where EBV plays an initiating role in oncogenesis but the viral genome is lost (70,71). Two recent studies identified “traces” of EBV infection in BL primary tumors where they detected EBV miRNA but not EBER (65,72). This raises the intriguing possibility that EBV− BL are actually derived from EBV-infected B cells which because of the absence of EBERs, a potential link with EBV can be missed. Analysis of cellular mutations in EBV− BL also has led to the hypothesis that EBV can be lost because compensatory mutations have occurred that substitute for the functions of EBV proteins (70). However, other studies that have identified unique gene expression profiles (GEPs) between EBV+ and EBV− BL (41) and evidence of ongoing somatic hypermutation in EBV+ BL but not in EBV− BL (39,73) which argue for different pathogenetic mechanisms driving the emergence of EBV+ and EBV− BL.
One possibility that has been addressed by a number of groups is whether there is a particular strain or variant of EBV associated with BL that had more oncogenic potential. However, the results have not been consistent and this question remains open. For example, Bhatia and colleagues classified five EBNA-1 subtypes and proposed that the V-leu subtype was more common in both BL as compared to healthy controls (74). However, a subsequent study by Rickinson and colleagues found no evidence of a selection of a particular EBNA-1 subtype in BL cases compared to controls (75). Similarly, Moormann and colleagues reported no evidence of a promoter variant, the ZpV3, in BL cases versus controls (76), while Kenney and colleagues did (77). Conversely, Grande et al. found no evidence of a bias towards EBV type 1 predominating in eBL (39) while Kaymaz et al. did (78). Earlier studies on HIV-associated and eBL found equal distribution of EBV types 1 and 2 (79) while BL cases in Brazil had a predominance of EBV type 1 (25). Unique variants of the LMP-1 have also been reported in eBL compared to controls (80,81). An important challenge in addressing this critical question is identifying the appropriate control population as a reference for the EBV genome strain(s) circulating within a population.
Because both strains of EBV are found in BL, an interesting question was whether there were cellular mutation differences in BL carrying either EBV type 1 or type 2. In one study, EBV type 2+ BL had less mutations in key genes compared to EBV type 1 (73,78). In contrast, Grande et al. (39) did not find genes that were differentially mutated in BL carrying either EBV type 1 and type 2 BL. Further studies on larger panels of BL tumors are required to resolve these discrepant results. It is possible that variations in EBV type between different geographic regions within sub-Saharan Africa could also account for these differences.
Activation-induced cytidine deaminase (AID) and BL
When the MYC translocation was first identified, there was speculation that it was mediated by the VDJ B cell recombinase used in the recombination of the variable and joining regions of the immunoglobulin genes (44). However, in 2008, Robbiani and colleagues provided the first evidence that the enzyme, activation induced cytidine deaminase (AID) (also called AICDA), was a cause of the MYC translocation (82). AID is the critical enzyme for somatic hypermutation and class switch recombination of immunoglobulin genes, key features in the generation of adaptive humoral immunity [reviewed in (83)]. AID induces point mutations by deaminating cytidine to uracil. Because of this, it is easy to identify cellular mutations as being caused by AID. AID can also induce DNA breaks in the immunoglobulin genes.
Because of the potential danger to the cell of having an enzyme capable of modifying the genome, AID is regulated not only at the transcriptional level, but also through translational, post-translational and sub-cellular localization (84). AID is also tightly regulated during B cell differentiation and expressed only in germinal center B cells in health (85). Evidence that both EBV and Plasmodium falciparum, etiologic co-factors in eBL, can aberrantly induce AID comes from both animal and cell culture models. Plasmodium infection alone can induce aberrant AID expression in B cells outside of the germinal center reaction in a mouse model of Plasmodium (86). Extracts from P. falciparum infected red blood cells were also shown to induce AID expression (RNA and protein) in tonsil B cells (87). More direct evidence of Plasmodium comes from a study by Robbiani et al. (88) where they repeatedly infected p53 deficient mice with Plasmodium chabaudi and induced lymphomas that had the characteristic MYC translocation on the IgH. Using an AID knockout mouse, they then showed that the capacity to induce a translocation was dependent on AID. Overexpression of MYC in normal B cells results in stress response apoptosis mediated by p53, ATM and p19 (89) suggesting that even if AID induces off-target translocation of MYC in normal cells, these cells will likely undergo apoptosis. Of note in the Robbiani study, no tumors occurred in mice in the absence of p53 implying that abrogation of apoptotic pathways is critical to emergence of the malignant cell in BL.
The story with EBV and AID activation is a bit more complicated. EBV infection of B cells ex vivo induces AID activity (90). Consistent with this, both the EBV LMP-1 as well as the EBNA-3C induce AID expression (91-93). AID was functional in these experiments as evidenced by accumulation of point mutations (91) as well as induction of T-cell independent class switching (92). LMP-1 induction of AID was indirect through upregulation of Egr-1 transcription factor (93). EBNA-3C was shown to bind directly to regulatory elements in the AICDA gene that encodes AID (91). However, overexpression of EBNA-2 inhibited AID expression (94) potentially counteracting LMP-1 and EBNA-3C induced AID activity. Moreover, EBV+ BL do not typically express LMP-1, EBNA-3C or EBNA-2 so if there is an EBV mediated effect on AID it preceedes oncogenesis.
Three studies in humans point to dysregulated AID expression in populations at risk for BL. First, Torgbor et al. (87) found AID frequently expressed in the tonsil lymphocytes of children from a malaria endemic region of Ghana but not in tonsil lymphocytes from children living in the US. In a study based in Kenya, elevated AID expression was correlated with peripheral EBV loads in children with repeated exposure to malaria (95). Elevated expression of AID in peripheral blood lymphocytes was detected prior to the emergence of BL in HIV+ patients (90) suggesting that dysregulation of AID is an early step prior to the MYC translocation. This is critical point as it leads to a hypothesis that chronic immune activation, regardless of inciting agent (e.g., viral or parasite) can drive the MYC translocation (96).
More recently, in a landmark paper, Staudt and colleagues sequenced DNA from over 100 EBV+ and EBV− BL biopsies and through analysis of both genome sequences and transcriptome data found that the genome wide mutational load was increased significantly in EBV+ BL compared to EBV− BL regardless of the epidemiology of the tumor (39). They hypothesized that the increased mutations were due to increased AID activity based on analysis of the point mutations that clearly have the mark of the cytidine deaminase. AID was also more highly expressed in eBL than in sBL. A subsequent study by Panea et al. (73) also found that EBV+ eBL had a mutational burden indicative of AID activity.
Other co-factors
When EBV was first discovered, it was thought (and hoped) that this would be the answer to understanding BL and possibly preventing BL. Unfortunately, EBV is a necessary but not sufficient cause of the malignant transformation that occurs in the EBV+ BL. As observed by Burkitt and colleagues early on, there was a clear epidemiologic link between cases of BL and endemic malaria transmission (97) and confirmed in later studies (7,98). However, it wasn’t until the 2000’s that we had a causal link both in the case control studies of Newton and colleagues in Malawi and Uganda where children with eBL had higher titers of anti-malaria antibodies as compared to controls (99,100). In addition, Robbiani by repeatedly infecting p53−/− mice with P. chaubadi, showed AID-dependent MYC translocated lymphoma similar to BL in humans (88). Combined, these studies provide the strongest evidence for a causal role of malaria in BL.
Within the context of both P. falciparum malaria and HIV, there is also a possible role for immunosuppression of EBV-specific responses. For example, Whittle et al. (101) demonstrated that peripheral blood lymphocyte (PBL) isolated from adult patients with acute malaria were unable to control outgrowth of EBV-transformed cells in a standard but relatively crude regression assay to assess EBV-specific T cell function. Moss et al. (102) demonstrated that healthy adults living in malaria holoendemic regions of Papua New Guinea had impaired EBV-specific T cells responses using the same regression assay. In children experiencing an episode of acute malaria, spontaneous outgrowth of EBV-transformed B cells ex vivo occurred at greater frequency (103). More recent studies show loss of EBV specific cytotoxic T lymphocyte (CTL) responses to lytic antigens in children living in a malaria holoendemic region (104) and a specific loss of IFNg CTL responses to EBNA-1 in eBL patients (105). A model put forward by Liu et al. (106) proposes that repeated infections with one pathogen (for example, Plasmodium) weakens the total CTL memory, such that established CTL memory to a different pathogen (such as EBV) will be diminished and ultimately collapse. This model could explain loss of EBV-immunity that has been reported in the above studies and could also account for how chronic antigen stimulation from not only P. falciparum but also other pathogens such as HIV could contribute to the emergence of BL.
If P. falciparum malaria and EBV act in concert to drive eBL, what are the factors that drive the other clinical variants of BL? One possibility is that factors that induce chronic activation of B cells and aberrant AID expression could trigger the MYC translocation. This model relies on B cell receptor (BCR) stimulation through extrinsic activation. Evidence of antigen selection of BCR in BL in both mouse models (107) and in humans (108) is found and suggests that other chronic infections including HIV could trigger lymphomagenesis (41,109). In contrast, analysis of BCR signaling in sBL is more indicative of mutations that allow tonic BCR signaling and not necessarily antigen selected (107,110). Because of the relatively low incidence of sBL, understanding the etiology becomes more challenging and has not been extensively explored. But it is clear that for all variants of BL, a MYC translocation and evasion of apoptosis are critical steps on the oncogenic pathway.
Defining BL based on the presence or absence of EBV
With the advent of more comprehensive “omic” techniques to evaluate the mutations in the cellular genome and define transcriptional profiles, these techniques have now been applied by several groups to identify molecular pathways that are disrupted in BL [(110-113), reviewed in (36)]. Common pathways dysregulated in BL include tonic B-cell receptor signaling, phosphoinositide 3-kinase (PI3K) pathway, and cyclin D3/CDK6 as well as mutations in the tumor suppressor, ID3, diminishing its function. Activating mutations in TCF3, a negative regulator of ID3 are also observed pointing to the critical importance of this pathway in driving cell proliferation. Other common mutations in BL include ARID1A and SMARCA4, part of the SWI/SNF family and involved in nucleosome remodeling. In addition, mutations are found in MCL1, FBX011, DDX3X, CCND3 as well as TP53 and MYC.
A challenge in compiling a comprehensive picture of the genomic landscape of BL is that while translocation of MYC and subsequent overexpression of MYC is the primary pathogenetic event, cellular mutations (e.g., deletions, amplifications, point mutations) evolve independently to target different cellular pathways (e.g., proliferation, apoptosis, immune escape). Genotype-phenotype correlations along with clinical correlations are needed to define the spectrum of mutations in BL and move into the field of precision medicine. And while, as noted above, certain patterns are emerging that shed light on the genetic basis of BL, it has becoming increasingly clear that the presence or absence of EBV in the lymphoma is the feature that defines other molecular differences (39). With this point in mind, it useful to go back and look at the reported differences in studies that compared eBL (almost always EBV+) and sBL (almost always EBV−) through the lens that EBV is the defining feature. Table 1 summarizes these studies and with this information a clear picture emerges of the molecular differences between EBV+ and EBV− BL.
Table 1
| Study | Analysis/sample type* | EBV+ | EBV− |
|---|---|---|---|
| Bellan |
Mutation rates in VH | 6.1 average mutation frequency in VH | 1.4 average mutation frequency in VH |
| Sequence analysis of CDR2 region for evidence of antigen selection | 11/18 evidence antigen selection | 0/13 evidence of antigen selection | |
| Piccaluga |
Gene expression profile (GEP) | 74 unique genes including BCR, TNFα/NF-κB, interleukin-dependent cascades | – |
| eBL |
↑miRNA | – | |
| EBV+ |
↑ |
– | |
| GC gene profile | GC gene profile | ||
| Navari |
GEP miRNA profile | Enrichment for metabolic processes | ↓27 miRNA |
| EBV+ |
↑has-miR-142-5p | ||
| Zhou |
Sanger sequencing of eBL and sBL, NGS of sBL | Mutations in |
Mutations in |
| eBL 100% EBV+ | Mutations in S22 and distal to AKT recognition | Mutations in T24 phosphorylation site | |
| Amato |
RNA seq/NGS sequencing of Ig genes | ↓Mutation frequency of TCF3/ID3 | ↑Mutation frequency of TCF3/ID3 |
| eBL |
Intraclonal diversity | ||
| Grande |
Whole genome sequencing, RNA-seq, miRNA-sea | ↑Mutation load per genome | ↓Mutation load |
| ↑Proportion of AID associated mutations | ↑TP53 mutations | ||
| ↑AID expression | – | ||
| Mutations in |
– | ||
| ↓Driver mutations esp. in apoptosis genes | – | ||
| ↑Noncoding mutations | ↑Coding mutations | ||
| Giulino-Roth |
NGS | Less likely to have genetic alterations | More likely to have multiple genetic alterations |
| Pediatric BL | |||
| Abate |
RNA seq | ↓Mutations in |
Mutations in MYC, ID3, TCF3, TP53 |
| eBL |
↑Mutations in |
TCF3 pathway more activated | |
| TCF pathway analysis on EBV+ |
|||
| Panea |
Whole genome sequencing, RNA seq | ↑Mutation load per genome | ↓Mutation load |
| ↑Proportion of AID associated mutations | |||
| ↑Mutations in |
|||
| ↓Mutations in |
|||
| Kaymaz |
RNAseq, gene set enrichment | 1,658 diff expressed genes | – |
| ↓ |
|||
| ↑DNA replication, mismatch repair, cell cycle regulation | |||
| Lenze |
miRNA microarray | ↑10/18 targets of miR-127 | – |
| eBL |
↑hsa-miR-10b, -216b, -499-3p | ↑hsa-miR-191, -374a, -193-5p |
AID, activation-induced cytidine deaminase; BCR, B cell receptor; BL, Burkitt lymphoma; eBL, endemic Burkitt lymphoma; EBV, Epstein-Barr virus; GEP, gene expression profile; ID-BL, immunodeficiency-associated Burkitt lymphoma; GC, germinal center; NGS, next-generation sequencing; sBL, sporadic Burkitt lymphoma.
The first key molecular difference in eBL vs. sBL was the breakpoint differences in the MYC translocation. In eBL, the translocation occurs 5’ of the MYC coding region while in sBL, the translocation typically occurs within the 1st intron or exon (20,117,118). Bellan and colleagues analyzed eBL, sBL and ID-BL for rearranged immunoglobulin heavy chain (VH) genes and compared them to germline VH segments (114). They found that the EBV+ BL regardless of epidemiologic origin, had more somatic mutations and evidence for antigen selection while the EBV− BL had only a small number of somatic mutations. Follow-up studies using next-generation sequencing (NGS) of Ig genes found intraclonal diversity supports a model for antigen-driven selection of BCRs in eBL (108). A subsequent GEP comparing eBL to sBL identified both similarities such that all BL were distinct from other non-Hodgkin lymphoma but also differences with 74 unique genes expressed in eBL but not sBL (41). An additional study evaluated EBV+ and EBV− ID-BL and again found differences, notably in 19 cellular miRNAs that were upregulated in the EBV+ BL compared to the EBV− BL (61). Both eBL and sBL had mutations in the forkhead box subtype (FOXO) transcription factor but there were differences in the presence of a mutation in the AKT recognition motif with T24 mutation common in sBL while the S22 was common in eBL (115). And as noted earlier both Grande and Panea identified evidence of AID mutation activity in EBV+ but it was not as extensive as in EBV− BL (39,73). In addition, Abate et al. (58) found evidence of decreased mutations in MYC, ID3, TCF3 and TP53 in EBV+ BL compared to EBV− BL while an increase in mutations in ARID1A, RHOA and CCNF. In addition, they also reported that the TCF3 target genes are more activated in EBV− BL (58). Interestingly, in EBV+ BL, noncoding mutations were in regulatory regions and likely due to AID activity (39). In this same study, they also found that mutations in apoptotic pathways defined EBV− BL but not EBV+ BL consistent with a hypothesis that one of EBV’s role in etiopathogenesis of BL is to abrogate apoptotic pathways.
An analysis of human miRNA in BL subtypes while finding differences in 38 miRNAs between BL and diffuse large B-cell lymphoma, also identified 6 miRNAs that were different between eBL and sBL (116). A subsequent study confirmed this observation that the differences were between EBV+ and EBV− BL and also found 10/18 targets of has miR-127 that were upregulated in EBV+ BL (41).
Since the discovery of EBV, the question has remained whether EBV is the driver or passenger on the road to malignant transformation of B-cells in BL. The theme emerging from the molecular analysis of BL is that the presence or absence of EBV is a defining feature of the clinical variant regardless of the epidemiology of the tumor. And that the convergence of similar pathology of clinical variants now shows that there are molecularly distinct EBV+ and EBV− entities. With this knowledge, we are still challenged to understand where in the B-cell differentiation pathway does EBV induce premalignant changes and increase susceptibility to malignant transformation.
In Figure 1, a model of etiologic pathways of EBV+ and EBV− BL is presented. In this model and consistent with pathologic findings, BL starts as a germinal center B cell, likely a centroblast. Both pathways posit 4 key features: (I) suppression of apoptosis, (II) activation of AID, (III) MYC translocation and (IV) accumulation of additional mutations that enhance cell proliferation/survival/growth. Where they differ is that EBV likely induces AID and accumulation of point mutations that distinguish the EBV+ and EBV− BL (39). Another key difference is the role of EBV in suppressing apoptotic pathways for EBV+ BL while mutations in ID3/TCF3/p53 play that role in EBV− BL (58). The steps in these two pathways remain to be fully elucidated.
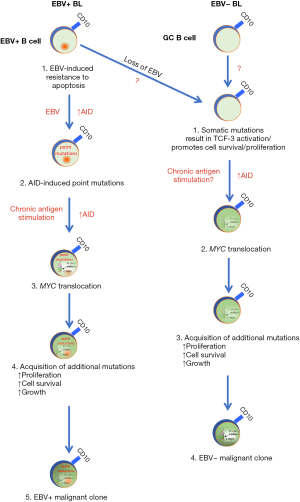
Conclusions
Guy de-Thé once called BL the Rosetta Stone of cancer (120). Through the study of this cancer first identified in Uganda, scientists have uncovered the first human cancer virus, developed the first human B cell line, identified the first human oncogene, and developed new models for cancer treatment that relied on chemotherapy alone. As we move further into the 21st century, BL is still serving as our guide as we uncover the mechanisms of oncogenesis and the differences in etiopathogenesis of EBV+ compared to EBV− BL.
Acknowledgments
This review is dedicated to the memory of Dr. Beverly Griffin whose passion and enthusiasm for studying Burkitt lymphoma is a reminder that there is still more work to be done while this cancer remains a lethal childhood cancer in sub-Saharan Africa.
Funding: This work was supported by the National Institutes of Health (AI141531).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Christopher P. Fox, Claire Shannon-Lowe) for the series “Lymphoma and Viruses” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/aol-21-18). The series “Lymphoma and Viruses” was commissioned by the editorial office without any funding or sponsorship. RR reports funding received from institution supported by NIH and that she served as a consultant for Viracta Therapeutics, Inc. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Burkitt D. A sarcoma involving the jaws in African children. Br J Surg 1958;46:218-23. [Crossref] [PubMed]
- Burkitt D. A children's cancer dependent on climatic factors. Nature 1962;194:232-4. [Crossref] [PubMed]
- Haddow AJ. An improved map for the study of Burkitt's lymphoma syndrome in Africa. East Afr Med J 1963;40:429-32. [PubMed]
- Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet 1964;1:702-3. [Crossref] [PubMed]
- Haddow AJ. Age incidence in Burkitt's lymphoma syndrome. East Afr Med J 1964;41:1-6. [PubMed]
- Hämmerl L, Colombet M, Rochford R, et al. The burden of Burkitt lymphoma in Africa. Infect Agent Cancer 2019;14:17. [Crossref] [PubMed]
- Rainey JJ, Mwanda WO, Wairiumu P, et al. Spatial distribution of Burkitt's lymphoma in Kenya and association with malaria risk. Trop Med Int Health 2007;12:936-43. [Crossref] [PubMed]
- Lavu E, Morewaya J, Maraka R, et al. Burkitt lymphoma in Papua New Guinea--40 years on. Ann Trop Paediatr 2005;25:191-7. [Crossref] [PubMed]
- Moss D. Epstein-Barr virus and malaria in relation to Burkitt's lymphoma in Papua New Guinea. P N G Med J 1986;29:41-3. [PubMed]
- Mwanda OW. Clinical characteristics of Burkitt's lymphoma seen in Kenyan patients. East Afr Med J 2004;S78-89. [Crossref] [PubMed]
- Offor UT, Akyea RK, Neequaye JE, et al. The changing clinical pattern of endemic Burkitt lymphoma in Western Africa: Experience from a tertiary center in Ghana. Pediatr Blood Cancer 2018;65:e27275 [Crossref] [PubMed]
- zur Hausen H, Schulte-Holthausen H, Klein G, et al. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 1970;228:1056-8. [Crossref] [PubMed]
- Shannon-Lowe C, Rickinson AB, Bell AI. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci 2017;372:20160271 [Crossref] [PubMed]
- Levine PH, Kamaraju LS, Connelly RR, et al. The American Burkitt's Lymphoma Registry: eight years' experience. Cancer 1982;49:1016-22. [Crossref] [PubMed]
- Satou A, Asano N, Nakazawa A, et al. Epstein-Barr virus (EBV)-positive sporadic burkitt lymphoma: an age-related lymphoproliferative disorder? Am J Surg Pathol 2015;39:227-35. [Crossref] [PubMed]
- Mbulaiteye SM, Anderson WF, Ferlay J, et al. Pediatric, elderly, and emerging adult-onset peaks in Burkitt's lymphoma incidence diagnosed in four continents, excluding Africa. Am J Hematol 2012;87:573-8. [Crossref] [PubMed]
- Mbulaiteye SM, Biggar RJ, Bhatia K, et al. Sporadic childhood Burkitt lymphoma incidence in the United States during 1992-2005. Pediatr Blood Cancer 2009;53:366-70. [Crossref] [PubMed]
- Ziegler JL. Burkitt's lymphoma. N Engl J Med 1981;305:735-45. [Crossref] [PubMed]
- Leoncini L, Raphael M, Stein H, et al. Burkitt lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al. editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edition. Lyon, France: IARC Press, 2008:262-4.
- Shiramizu B, Barriga F, Neequaye J, et al. Patterns of chromosomal breakpoint locations in Burkitt's lymphoma: relevance to geography and Epstein-Barr virus association. Blood 1991;77:1516-26. [Crossref] [PubMed]
- Guech-Ongey M, Simard EP, Anderson WF, et al. AIDS-related Burkitt lymphoma in the United States: what do age and CD4 lymphocyte patterns tell us about etiology and/or biology? Blood 2010;116:5600-4. [Crossref] [PubMed]
- Bellan C, Lazzi S, De Falco G, et al. Burkitt's lymphoma: new insights into molecular pathogenesis. J Clin Pathol 2003;56:188-92. [Crossref] [PubMed]
- Kimani SM, Painschab MS, Horner MJ, et al. Epidemiology of haematological malignancies in people living with HIV. Lancet HIV 2020;7:e641-51. [Crossref] [PubMed]
- Mbulaiteye SM, Pullarkat ST, Nathwani BN, et al. Epstein-Barr virus patterns in US Burkitt lymphoma tumors from the SEER residual tissue repository during 1979-2009. APMIS 2014;122:5-15. [Crossref] [PubMed]
- Queiroga EM, Gualco G, Weiss LM, et al. Burkitt lymphoma in Brazil is characterized by geographically distinct clinicopathologic features. Am J Clin Pathol 2008;130:946-56. [Crossref] [PubMed]
- Hunt BJ, Thomas JA, Burke M, et al. Epstein-Barr virus associated Burkitt lymphoma in a heart transplant recipient. Transplantation 1996;62:869-72. [Crossref] [PubMed]
- Akar Özkan E, Özdemir BH, Akdur A, et al. Burkitt lymphoma after transplant: an aggressive lymphoproliferative disease. Exp Clin Transplant 2014;12:136-8. [PubMed]
- Picarsic J, Jaffe R, Mazariegos G, et al. Post-transplant Burkitt lymphoma is a more aggressive and distinct form of post-transplant lymphoproliferative disorder. Cancer 2011;117:4540-50. [Crossref] [PubMed]
- Mbulaiteye SM, Clarke CA, Morton LM, et al. Burkitt lymphoma risk in U.S. solid organ transplant recipients. Am J Hematol 2013;88:245-50. [Crossref] [PubMed]
- Gutiérrez MI, Bhatia K, Barriga F, et al. Molecular epidemiology of Burkitt's lymphoma from South America: differences in breakpoint location and Epstein-Barr virus association from tumors in other world regions. Blood 1992;79:3261-6. [Crossref] [PubMed]
- Bacchi MM, Bacchi CE, Alvarenga M, et al. Burkitt's lymphoma in Brazil: strong association with Epstein-Barr virus. Mod Pathol 1996;9:63-7. [PubMed]
- Klumb CE, Hassan R, De Oliveira DE, et al. Geographic variation in Epstein-Barr virus-associated Burkitt's lymphoma in children from Brazil. Int J Cancer 2004;108:66-70. [Crossref] [PubMed]
- Hassan R, Klumb CE, Felisbino FE, et al. Clinical and demographic characteristics of Epstein-Barr virus-associated childhood Burkitt's lymphoma in Southeastern Brazil: epidemiological insights from an intermediate risk region. Haematologica 2008;93:780-3. [Crossref] [PubMed]
- Lara J, Cohen M, De Matteo E, et al. Epstein-Barr virus (EBV) association and latency profile in pediatric Burkitt's lymphoma: experience of a single institution in Argentina. J Med Virol 2014;86:845-50. [Crossref] [PubMed]
- Dojcinov SD, Fend F, Quintanilla-Martinez L. EBV-Positive Lymphoproliferations of B- T- and NK-Cell Derivation in Non-Immunocompromised Hosts. Pathogens 2018;7:28. [Crossref] [PubMed]
- Schmitz R, Ceribelli M, Pittaluga S, et al. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med 2014;4:a014282 [Crossref] [PubMed]
- Wright DH. Burkitt's lymphoma: a review of the pathology, immunology, and possible etiologic factors. Pathol Annu 1971;6:337-63. [PubMed]
- Ok CY, Medeiros LJ. High-grade B-cell lymphoma: a term re-purposed in the revised WHO classification. Pathology 2020;52:68-77. [Crossref] [PubMed]
- Grande BM, Gerhard DS, Jiang A, et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019;133:1313-24. [Crossref] [PubMed]
- Dogan A, Bagdi E, Munson P, et al. CD10 and BCL-6 expression in paraffin sections of normal lymphoid tissue and B-cell lymphomas. Am J Surg Pathol 2000;24:846-52. [Crossref] [PubMed]
- Piccaluga PP, De Falco G, Kustagi M, et al. Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood 2011;117:3596-608. [Crossref] [PubMed]
- Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med 2006;354:2431-42. [Crossref] [PubMed]
- Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med 2006;354:2419-30. [Crossref] [PubMed]
- Magrath I. The pathogenesis of Burkitt's lymphoma. Adv Cancer Res 1990;55:133-270. [Crossref] [PubMed]
- Swerdlow SH, Campo E, Harris NL, et al. editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edition. Lyon, France: IARC Press, 2008.
- López C, Kleinheinz K, Aukema SM, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun 2019;10:1459. [Crossref] [PubMed]
- Brady G, MacArthur GJ, Farrell PJ. Epstein-Barr virus and Burkitt lymphoma. J Clin Pathol 2007;60:1397-402. [PubMed]
- Neri A, Barriga F, Inghirami G, et al. Epstein-Barr virus infection precedes clonal expansion in Burkitt's and acquired immunodeficiency syndrome-associated lymphoma. Blood 1991;77:1092-5. [Crossref] [PubMed]
- Geser A, de Thé G, Lenoir G, et al. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt's lymphoma. Int J Cancer 1982;29:397-400. [Crossref] [PubMed]
- Kennedy G, Komano J, Sugden B. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc Natl Acad Sci U S A 2003;100:14269-74. [Crossref] [PubMed]
- Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, et al. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol 1994;68:6069-73. [Crossref] [PubMed]
- Kang MS, Kieff E. Epstein-Barr virus latent genes. Exp Mol Med 2015;47:e131 [Crossref] [PubMed]
- Rowe M, Rowe DT, Gregory CD, et al. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J 1987;6:2743-51. [Crossref] [PubMed]
- Wilson JB, Bell JL, Levine AJ. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J 1996;15:3117-26. [Crossref] [PubMed]
- Kang MS, Soni V, Bronson R, et al. Epstein-Barr virus nuclear antigen 1 does not cause lymphoma in C57BL/6J mice. J Virol 2008;82:4180-3. [Crossref] [PubMed]
- Holowaty MN, Frappier L. HAUSP/USP7 as an Epstein-Barr virus target. Biochem Soc Trans 2004;32:731-2. [Crossref] [PubMed]
- Xue SA, Labrecque LG, Lu QL, et al. Promiscuous expression of Epstein-Barr virus genes in Burkitt's lymphoma from the central African country Malawi. Int J Cancer 2002;99:635-43. [Crossref] [PubMed]
- Abate F, Ambrosio MR, Mundo L, et al. Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma. PLoS Pathog 2015;11:e1005158 [Crossref] [PubMed]
- Komano J, Maruo S, Kurozumi K, et al. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol 1999;73:9827-31. [Crossref] [PubMed]
- Ambrosio MR, Navari M, Di Lisio L, et al. The Epstein Barr-encoded BART-6-3p microRNA affects regulation of cell growth and immuno response in Burkitt lymphoma. Infect Agent Cancer 2014;9:12. [Crossref] [PubMed]
- Navari M, Fuligni F, Laginestra MA, et al. Molecular signature of Epstein Barr virus-positive Burkitt lymphoma and post-transplant lymphoproliferative disorder suggest different roles for Epstein Barr virus. Front Microbiol 2014;5:728. [Crossref] [PubMed]
- Xia T, O'Hara A, Araujo I, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res 2008;68:1436-42. [Crossref] [PubMed]
- Navari M, Etebari M, De Falco G, et al. The presence of Epstein-Barr virus significantly impacts the transcriptional profile in immunodeficiency-associated Burkitt lymphoma. Front Microbiol 2015;6:556. [Crossref] [PubMed]
- Vereide DT, Seto E, Chiu YF, et al. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 2014;33:1258-64. [Crossref] [PubMed]
- Mundo L, Ambrosio MR, Picciolini M, et al. Unveiling Another Missing Piece in EBV-Driven Lymphomagenesis: EBV-Encoded MicroRNAs Expression in EBER-Negative Burkitt Lymphoma Cases. Front Microbiol 2017;8:229. [Crossref] [PubMed]
- Niedobitek G, Agathanggelou A, Rowe M, et al. Heterogeneous expression of Epstein-Barr virus latent proteins in endemic Burkitt's lymphoma. Blood 1995;86:659-65. [Crossref] [PubMed]
- Labrecque LG, Xue SA, Kazembe P, et al. Expression of Epstein-Barr virus lytically related genes in African Burkitt's lymphoma: correlation with patient response to therapy. Int J Cancer 1999;81:6-11. [Crossref] [PubMed]
- Kelly G, Bell A, Rickinson A. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med 2002;8:1098-104. [Crossref] [PubMed]
- Allday MJ. How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt's lymphoma? Semin Cancer Biol 2009;19:366-76. [Crossref] [PubMed]
- Hutcheson RL, Chakravorty A, Sugden B. Burkitt Lymphomas Evolve to Escape Dependencies on Epstein-Barr Virus. Front Cell Infect Microbiol 2021;10:606412 [Crossref] [PubMed]
- Razzouk BI, Srinivas S, Sample CE, et al. Epstein-Barr Virus DNA recombination and loss in sporadic Burkitt's lymphoma. J Infect Dis 1996;173:529-35. [Crossref] [PubMed]
- Mundo L, Del Porro L, Granai M, et al. Frequent traces of EBV infection in Hodgkin and non-Hodgkin lymphomas classified as EBV-negative by routine methods: expanding the landscape of EBV-related lymphomas. Mod Pathol 2020;33:2407-21. [Crossref] [PubMed]
- Panea RI, Love CL, Shingleton JR, et al. The whole-genome landscape of Burkitt lymphoma subtypes. Blood 2019;134:1598-607. [Crossref] [PubMed]
- Gutiérrez MI, Raj A, Spangler G, et al. Sequence variations in EBNA-1 may dictate restriction of tissue distribution of Epstein-Barr virus in normal and tumour cells. J Gen Virol 1997;78:1663-70. [Crossref] [PubMed]
- Habeshaw G, Yao QY, Bell AI, et al. Epstein-barr virus nuclear antigen 1 sequences in endemic and sporadic Burkitt's lymphoma reflect virus strains prevalent in different geographic areas. J Virol 1999;73:965-75. [Crossref] [PubMed]
- Kaymaz Y, Oduor CI, Aydemir O, et al. Epstein-Barr Virus Genomes Reveal Population Structure and Type 1 Association with Endemic Burkitt Lymphoma. J Virol 2020;94:e02007-19. [Crossref] [PubMed]
- Bristol JA, Djavadian R, Albright ER, et al. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog 2018;14:e1007179 [Crossref] [PubMed]
- Kaymaz Y, Oduor CI, Yu H, et al. Comprehensive Transcriptome and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV Type-Specific Differences. Mol Cancer Res 2017;15:563-76. [Crossref] [PubMed]
- Goldschmidts WL, Bhatia K, Johnson JF, et al. Epstein-Barr virus genotypes in AIDS-associated lymphomas are similar to those in endemic Burkitt's lymphomas. Leukemia 1992;6:875-8. [PubMed]
- Wohlford EM, Asito AS, Chelimo K, et al. Identification of a novel variant of LMP-1 of EBV in patients with endemic Burkitt lymphoma in western Kenya. Infect Agent Cancer 2013;8:34. [Crossref] [PubMed]
- Liao HM, Liu H, Lei H, et al. Frequency of EBV LMP-1 Promoter and Coding Variations in Burkitt Lymphoma Samples in Africa and South America and Peripheral Blood in Uganda. Cancers (Basel) 2018;10:177. [Crossref] [PubMed]
- Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 2008;135:1028-38. [Crossref] [PubMed]
- Feng Y, Seija N, Di Noia JM, et al. AID in Antibody Diversification: There and Back Again. Trends Immunol 2020;41:586-600. [Crossref] [PubMed]
- Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity 2013;46:83-101. [Crossref] [PubMed]
- Huang C. Germinal Center Reaction. Adv Exp Med Biol 2020;1254:47-53. [Crossref] [PubMed]
- Wilmore JR, Maue AC, Rochford R. Plasmodium chabaudi infection induces AID expression in transitional and marginal zone B cells. Immun Inflamm Dis 2016;4:497-505. [Crossref] [PubMed]
- Torgbor C, Awuah P, Deitsch K, et al. A multifactorial role for P. falciparum malaria in endemic Burkitt's lymphoma pathogenesis. PLoS Pathog 2014;10:e1004170 [Crossref] [PubMed]
- Robbiani DF, Deroubaix S, Feldhahn N, et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell 2015;162:727-37. [Crossref] [PubMed]
- Jankovic M, Robbiani DF, Dorsett Y, et al. Role of the translocation partner in protection against AID-dependent chromosomal translocations. Proc Natl Acad Sci U S A 2010;107:187-92. [Crossref] [PubMed]
- Epeldegui M, Breen EC, Hung YP, et al. Elevated expression of activation induced cytidine deaminase in peripheral blood mononuclear cells precedes AIDS-NHL diagnosis. AIDS 2007;21:2265-70. [Crossref] [PubMed]
- Kalchschmidt JS, Bashford-Rogers R, Paschos K, et al. Epstein-Barr virus nuclear protein EBNA3C directly induces expression of AID and somatic mutations in B cells. J Exp Med 2016;213:921-8. [Crossref] [PubMed]
- He B, Raab-Traub N, Casali P, et al. EBV-encoded latent membrane protein 1 cooperates with BAFF/BLyS and APRIL to induce T cell-independent Ig heavy chain class switching. J Immunol 2003;171:5215-24. [Crossref] [PubMed]
- Kim JH, Kim WS, Park C. Epstein-Barr virus latent membrane protein 1 increases genomic instability through Egr-1-mediated up-regulation of activation-induced cytidine deaminase in B-cell lymphoma. Leuk Lymphoma 2013;54:2035-40. [Crossref] [PubMed]
- Tobollik S, Meyer L, Buettner M, et al. Epstein-Barr virus nuclear antigen 2 inhibits AID expression during EBV-driven B-cell growth. Blood 2006;108:3859-64. [Crossref] [PubMed]
- Wilmore JR, Asito AS, Wei C, et al. AID expression in peripheral blood of children living in a malaria holoendemic region is associated with changes in B cell subsets and Epstein-Barr virus. Int J Cancer 2015;136:1371-80. [Crossref] [PubMed]
- Epeldegui M, Martínez-Maza O. Immune Activation: Contribution to AIDS-Associated Non-Hodgkin Lymphoma. For Immunopathol Dis Therap 2015;6:79-90. [Crossref] [PubMed]
- . BURKITT D. Determining the climatic limitations of a children's cancer common in Africa. Br Med J 1962;2:1019-23. [Crossref] [PubMed]
- Morrow RH. Epidemiological evidence for the role of faciparum malaria in the pathogenesis of Burkitt's lymphoma. In: Lenoir GM, O'Conor GT, Olweny CLM. editors. Burkitt's Lymphoma: a human cancer model. IARC Scientific Publication No. 60. Lyon: IARC Press, 1985:177-85.
- Carpenter LM, Newton R, Casabonne D, et al. Antibodies against malaria and Epstein-Barr virus in childhood Burkitt lymphoma: a case-control study in Uganda. Int J Cancer 2008;122:1319-23. [Crossref] [PubMed]
- Mutalima N, Molyneux E, Jaffe H, et al. Associations between Burkitt lymphoma among children in Malawi and infection with HIV, EBV and malaria: results from a case-control study. PLoS One 2008;3:e2505 [Crossref] [PubMed]
- Whittle HC, Brown J, Marsh K, et al. T-cell control of Epstein-Barr virus-infected B cells is lost during P. falciparum malaria. Nature 1984;312:449-50. [Crossref] [PubMed]
- Moss DJ, Burrows SR, Castelino DJ, et al. A comparison of Epstein-Barr virus-specific T-cell immunity in malaria-endemic and -nonendemic regions of Papua New Guinea. Int J Cancer 1983;31:727-32. [Crossref] [PubMed]
- Whittle HC, Brown J, Marsh K, et al. The effects of Plasmodium falciparum malaria on immune control of B lymphocytes in Gambian children. Clin Exp Immunol 1990;80:213-8. [Crossref] [PubMed]
- Moormann AM, Chelimo K, Sumba PO, et al. Exposure to holoendemic malaria results in suppression of Epstein-Barr virus-specific T cell immunosurveillance in Kenyan children. J Infect Dis 2007;195:799-808. [Crossref] [PubMed]
- Moormann AM, Heller KN, Chelimo K, et al. Children with endemic Burkitt lymphoma are deficient in EBNA1-specific IFN-gamma T cell responses. Int J Cancer 2009;124:1721-6. [Crossref] [PubMed]
- Liu H, Andreansky S, Diaz G, et al. Quantitative analysis of long-term virus-specific CD8+-T-cell memory in mice challenged with unrelated pathogens. J Virol 2003;77:7756-63. [Crossref] [PubMed]
- Refaeli Y, Young RM, Turner BC, et al. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol 2008;6:e152 [Crossref] [PubMed]
- Amato T, Abate F, Piccaluga P, et al. Clonality Analysis of Immunoglobulin Gene Rearrangement by Next-Generation Sequencing in Endemic Burkitt Lymphoma Suggests Antigen Drive Activation of BCR as Opposed to Sporadic Burkitt Lymphoma. Am J Clin Pathol 2016;145:116-27. [Crossref] [PubMed]
- Capello D, Martini M, Gloghini A, et al. Molecular analysis of immunoglobulin variable genes in human immunodeficiency virus-related non-Hodgkin's lymphoma reveals implications for disease pathogenesis and histogenesis. Haematologica 2008;93:1178-85. [Crossref] [PubMed]
- Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 2012;490:116-20. [Crossref] [PubMed]
- Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet 2012;44:1316-20. [Crossref] [PubMed]
- Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 2012;44:1321-5. [Crossref] [PubMed]
- Giulino-Roth L, Wang K, MacDonald TY, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood 2012;120:5181-4. [Crossref] [PubMed]
- Bellan C, Lazzi S, Hummel M, et al. Immunoglobulin gene analysis reveals 2 distinct cells of origin for EBV-positive and EBV-negative Burkitt lymphomas. Blood 2005;106:1031-6. [Crossref] [PubMed]
- Zhou P, Blain AE, Newman AM, et al. Sporadic and endemic Burkitt lymphoma have frequent FOXO1 mutations but distinct hotspots in the AKT recognition motif. Blood Adv 2019;3:2118-27. [Crossref] [PubMed]
- Lenze D, Leoncini L, Hummel M, et al. The different epidemiologic subtypes of Burkitt lymphoma share a homogenous micro RNA profile distinct from diffuse large B-cell lymphoma. Leukemia 2011;25:1869-76. [Crossref] [PubMed]
- Neri A, Barriga F, Knowles DM, et al. Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A 1988;85:2748-52. [Crossref] [PubMed]
- Pelicci PG, Knowles DM 2nd, Magrath I, et al. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A 1986;83:2984-8. [Crossref] [PubMed]
- Paschos K, Smith P, Anderton E, et al. Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog 2009;5:e1000492 [Crossref] [PubMed]
- de-Thé G. The Epstein-Barr virus (EBV): a Rosetta Stone for understanding the role of viruses in immunopathological disorders and in human carcinogenesis. Biomed Pharmacother 1985;39:49-51. [PubMed]
Cite this article as: Rochford R. Reframing Burkitt lymphoma: virology not epidemiology defines clinical variants. Ann Lymphoma 2021;5:22.


